
Aquimarina brevivitae
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Aquimarina
Average proteome isoelectric point is 6.47
Get precalculated fractions of proteins
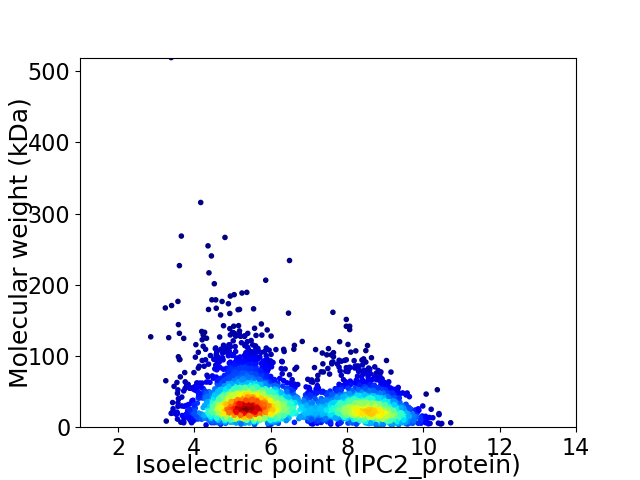
Virtual 2D-PAGE plot for 3416 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4Q7PGE2|A0A4Q7PGE2_9FLAO Cytochrome c peroxidase OS=Aquimarina brevivitae OX=323412 GN=EV197_0783 PE=4 SV=1
MM1 pKa = 7.91KK2 pKa = 9.85YY3 pKa = 10.59LKK5 pKa = 10.26IIIGLFLSLTVIVSCKK21 pKa = 9.99EE22 pKa = 3.81DD23 pKa = 3.78DD24 pKa = 3.61SFEE27 pKa = 4.12YY28 pKa = 10.74LDD30 pKa = 4.51AVEE33 pKa = 4.77APSNITALVNITQDD47 pKa = 2.77NTGFVTIIPTGKK59 pKa = 9.76GAVSFDD65 pKa = 3.65VYY67 pKa = 10.97FGDD70 pKa = 4.16GTLEE74 pKa = 4.14PVNVTPGEE82 pKa = 4.22KK83 pKa = 9.71AEE85 pKa = 4.7HH86 pKa = 6.53IYY88 pKa = 11.08EE89 pKa = 4.37EE90 pKa = 4.49GSYY93 pKa = 9.62EE94 pKa = 3.73LRR96 pKa = 11.84IVGKK100 pKa = 9.71SVSGKK105 pKa = 4.98TTEE108 pKa = 3.88AVQPLVVSFNPPQNLEE124 pKa = 3.96VLIEE128 pKa = 3.94NDD130 pKa = 3.25AAVSKK135 pKa = 10.16QVNVTATAEE144 pKa = 4.14FAVSYY149 pKa = 10.57DD150 pKa = 3.47VYY152 pKa = 10.87FGEE155 pKa = 4.7NADD158 pKa = 4.13DD159 pKa = 4.12TPVSANIGDD168 pKa = 4.04TVSYY172 pKa = 10.0IYY174 pKa = 10.71EE175 pKa = 3.93NAGTYY180 pKa = 7.54TIRR183 pKa = 11.84VVARR187 pKa = 11.84GGAIEE192 pKa = 4.09TTEE195 pKa = 3.93YY196 pKa = 10.73TEE198 pKa = 4.21EE199 pKa = 4.09FTVTEE204 pKa = 3.87ILQPVASAPAPPARR218 pKa = 11.84STQDD222 pKa = 3.19VISIFSAAYY231 pKa = 10.21DD232 pKa = 3.79DD233 pKa = 4.86VAGTDD238 pKa = 3.87YY239 pKa = 11.09FPDD242 pKa = 3.45WGQGSQGSSWSSFDD256 pKa = 4.49LNGDD260 pKa = 3.55EE261 pKa = 3.92MLQYY265 pKa = 10.96INLSYY270 pKa = 10.74QGIQFGSPVDD280 pKa = 3.64VSNMEE285 pKa = 4.85FIHH288 pKa = 7.55LDD290 pKa = 2.85VWTSGDD296 pKa = 3.14VSRR299 pKa = 11.84IEE301 pKa = 3.83TSLISVSNGEE311 pKa = 4.02RR312 pKa = 11.84PVWSDD317 pKa = 2.67LTAGEE322 pKa = 4.39WTSIDD327 pKa = 3.6IPISAYY333 pKa = 9.27TDD335 pKa = 3.29QNGFTVADD343 pKa = 3.31IHH345 pKa = 5.79QLKK348 pKa = 10.13FVGDD352 pKa = 3.92PWAAGTVFIDD362 pKa = 3.47NIYY365 pKa = 10.25FYY367 pKa = 10.59RR368 pKa = 11.84APTEE372 pKa = 4.23VINFPINFEE381 pKa = 4.79SNNLTYY387 pKa = 9.62TWSGFGNADD396 pKa = 4.33FGPIPASTVNNPDD409 pKa = 3.36PSGMNFSNTVVEE421 pKa = 4.2IQKK424 pKa = 10.0PAGAQVWAGASMNLSGTADD443 pKa = 4.23FSNGTTVTMKK453 pKa = 9.95VWSPRR458 pKa = 11.84AGVPVLFKK466 pKa = 10.68MEE468 pKa = 4.66DD469 pKa = 3.25QTSAPDD475 pKa = 3.73GNGNPSVFVEE485 pKa = 4.57VQATTTVANAWEE497 pKa = 4.32EE498 pKa = 4.02LSFDD502 pKa = 3.46LTSYY506 pKa = 11.19NGFSTSIPYY515 pKa = 10.69SNVIIFPDD523 pKa = 4.56FGNMGAGEE531 pKa = 4.55AFYY534 pKa = 10.84FDD536 pKa = 5.54DD537 pKa = 5.8IEE539 pKa = 4.26IASLKK544 pKa = 10.6LPLNFEE550 pKa = 4.44VSTLTYY556 pKa = 8.48TWAGFGDD563 pKa = 4.57PNFGPIPAATATNPDD578 pKa = 3.24QTGLNTSATVTEE590 pKa = 4.26IQKK593 pKa = 8.85PTGSQVWAGASLALDD608 pKa = 3.96GPVDD612 pKa = 4.25FSNGTTISLKK622 pKa = 9.73VWSPRR627 pKa = 11.84AGVPILFKK635 pKa = 10.6MEE637 pKa = 4.41DD638 pKa = 3.49TTSPPDD644 pKa = 3.51GNGNPTVFVEE654 pKa = 4.68VQSSTTVANSWEE666 pKa = 4.66EE667 pKa = 3.6ITFDD671 pKa = 3.27LTSFNGFSTDD681 pKa = 2.37ISYY684 pKa = 11.51YY685 pKa = 10.52NVIIFPDD692 pKa = 4.59FGNMGAGEE700 pKa = 4.45TFYY703 pKa = 11.13FDD705 pKa = 7.19DD706 pKa = 5.0IFLTNN711 pKa = 3.53
MM1 pKa = 7.91KK2 pKa = 9.85YY3 pKa = 10.59LKK5 pKa = 10.26IIIGLFLSLTVIVSCKK21 pKa = 9.99EE22 pKa = 3.81DD23 pKa = 3.78DD24 pKa = 3.61SFEE27 pKa = 4.12YY28 pKa = 10.74LDD30 pKa = 4.51AVEE33 pKa = 4.77APSNITALVNITQDD47 pKa = 2.77NTGFVTIIPTGKK59 pKa = 9.76GAVSFDD65 pKa = 3.65VYY67 pKa = 10.97FGDD70 pKa = 4.16GTLEE74 pKa = 4.14PVNVTPGEE82 pKa = 4.22KK83 pKa = 9.71AEE85 pKa = 4.7HH86 pKa = 6.53IYY88 pKa = 11.08EE89 pKa = 4.37EE90 pKa = 4.49GSYY93 pKa = 9.62EE94 pKa = 3.73LRR96 pKa = 11.84IVGKK100 pKa = 9.71SVSGKK105 pKa = 4.98TTEE108 pKa = 3.88AVQPLVVSFNPPQNLEE124 pKa = 3.96VLIEE128 pKa = 3.94NDD130 pKa = 3.25AAVSKK135 pKa = 10.16QVNVTATAEE144 pKa = 4.14FAVSYY149 pKa = 10.57DD150 pKa = 3.47VYY152 pKa = 10.87FGEE155 pKa = 4.7NADD158 pKa = 4.13DD159 pKa = 4.12TPVSANIGDD168 pKa = 4.04TVSYY172 pKa = 10.0IYY174 pKa = 10.71EE175 pKa = 3.93NAGTYY180 pKa = 7.54TIRR183 pKa = 11.84VVARR187 pKa = 11.84GGAIEE192 pKa = 4.09TTEE195 pKa = 3.93YY196 pKa = 10.73TEE198 pKa = 4.21EE199 pKa = 4.09FTVTEE204 pKa = 3.87ILQPVASAPAPPARR218 pKa = 11.84STQDD222 pKa = 3.19VISIFSAAYY231 pKa = 10.21DD232 pKa = 3.79DD233 pKa = 4.86VAGTDD238 pKa = 3.87YY239 pKa = 11.09FPDD242 pKa = 3.45WGQGSQGSSWSSFDD256 pKa = 4.49LNGDD260 pKa = 3.55EE261 pKa = 3.92MLQYY265 pKa = 10.96INLSYY270 pKa = 10.74QGIQFGSPVDD280 pKa = 3.64VSNMEE285 pKa = 4.85FIHH288 pKa = 7.55LDD290 pKa = 2.85VWTSGDD296 pKa = 3.14VSRR299 pKa = 11.84IEE301 pKa = 3.83TSLISVSNGEE311 pKa = 4.02RR312 pKa = 11.84PVWSDD317 pKa = 2.67LTAGEE322 pKa = 4.39WTSIDD327 pKa = 3.6IPISAYY333 pKa = 9.27TDD335 pKa = 3.29QNGFTVADD343 pKa = 3.31IHH345 pKa = 5.79QLKK348 pKa = 10.13FVGDD352 pKa = 3.92PWAAGTVFIDD362 pKa = 3.47NIYY365 pKa = 10.25FYY367 pKa = 10.59RR368 pKa = 11.84APTEE372 pKa = 4.23VINFPINFEE381 pKa = 4.79SNNLTYY387 pKa = 9.62TWSGFGNADD396 pKa = 4.33FGPIPASTVNNPDD409 pKa = 3.36PSGMNFSNTVVEE421 pKa = 4.2IQKK424 pKa = 10.0PAGAQVWAGASMNLSGTADD443 pKa = 4.23FSNGTTVTMKK453 pKa = 9.95VWSPRR458 pKa = 11.84AGVPVLFKK466 pKa = 10.68MEE468 pKa = 4.66DD469 pKa = 3.25QTSAPDD475 pKa = 3.73GNGNPSVFVEE485 pKa = 4.57VQATTTVANAWEE497 pKa = 4.32EE498 pKa = 4.02LSFDD502 pKa = 3.46LTSYY506 pKa = 11.19NGFSTSIPYY515 pKa = 10.69SNVIIFPDD523 pKa = 4.56FGNMGAGEE531 pKa = 4.55AFYY534 pKa = 10.84FDD536 pKa = 5.54DD537 pKa = 5.8IEE539 pKa = 4.26IASLKK544 pKa = 10.6LPLNFEE550 pKa = 4.44VSTLTYY556 pKa = 8.48TWAGFGDD563 pKa = 4.57PNFGPIPAATATNPDD578 pKa = 3.24QTGLNTSATVTEE590 pKa = 4.26IQKK593 pKa = 8.85PTGSQVWAGASLALDD608 pKa = 3.96GPVDD612 pKa = 4.25FSNGTTISLKK622 pKa = 9.73VWSPRR627 pKa = 11.84AGVPILFKK635 pKa = 10.6MEE637 pKa = 4.41DD638 pKa = 3.49TTSPPDD644 pKa = 3.51GNGNPTVFVEE654 pKa = 4.68VQSSTTVANSWEE666 pKa = 4.66EE667 pKa = 3.6ITFDD671 pKa = 3.27LTSFNGFSTDD681 pKa = 2.37ISYY684 pKa = 11.51YY685 pKa = 10.52NVIIFPDD692 pKa = 4.59FGNMGAGEE700 pKa = 4.45TFYY703 pKa = 11.13FDD705 pKa = 7.19DD706 pKa = 5.0IFLTNN711 pKa = 3.53
Molecular weight: 76.73 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Q7P1B5|A0A4Q7P1B5_9FLAO SprB-like repeat protein (Fragment) OS=Aquimarina brevivitae OX=323412 GN=EV197_2570 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.59RR3 pKa = 11.84TFQPSKK9 pKa = 10.15RR10 pKa = 11.84KK11 pKa = 9.6RR12 pKa = 11.84KK13 pKa = 8.29NKK15 pKa = 9.34HH16 pKa = 4.03GFRR19 pKa = 11.84EE20 pKa = 4.05RR21 pKa = 11.84MASVNGRR28 pKa = 11.84KK29 pKa = 9.21VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.09GRR39 pKa = 11.84KK40 pKa = 7.97KK41 pKa = 10.59LSVSSEE47 pKa = 3.56IRR49 pKa = 11.84HH50 pKa = 4.99KK51 pKa = 10.53HH52 pKa = 4.45
MM1 pKa = 7.45KK2 pKa = 9.59RR3 pKa = 11.84TFQPSKK9 pKa = 10.15RR10 pKa = 11.84KK11 pKa = 9.6RR12 pKa = 11.84KK13 pKa = 8.29NKK15 pKa = 9.34HH16 pKa = 4.03GFRR19 pKa = 11.84EE20 pKa = 4.05RR21 pKa = 11.84MASVNGRR28 pKa = 11.84KK29 pKa = 9.21VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.09GRR39 pKa = 11.84KK40 pKa = 7.97KK41 pKa = 10.59LSVSSEE47 pKa = 3.56IRR49 pKa = 11.84HH50 pKa = 4.99KK51 pKa = 10.53HH52 pKa = 4.45
Molecular weight: 6.26 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1162327 |
27 |
4967 |
340.3 |
38.38 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.545 ± 0.045 | 0.751 ± 0.015 |
5.736 ± 0.034 | 6.59 ± 0.042 |
5.007 ± 0.033 | 6.298 ± 0.047 |
1.725 ± 0.021 | 7.891 ± 0.038 |
7.493 ± 0.065 | 9.374 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.01 ± 0.022 | 5.892 ± 0.045 |
3.453 ± 0.026 | 3.785 ± 0.024 |
3.492 ± 0.033 | 6.323 ± 0.035 |
6.151 ± 0.059 | 6.206 ± 0.033 |
1.037 ± 0.013 | 4.242 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
