
Heterocapsa circularisquama RNA virus 01 (strain 34) (HcRNAV01)
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Sobelivirales; Alvernaviridae; Dinornavirus; Heterocapsa circularisquama RNA virus 01
Average proteome isoelectric point is 7.63
Get precalculated fractions of proteins
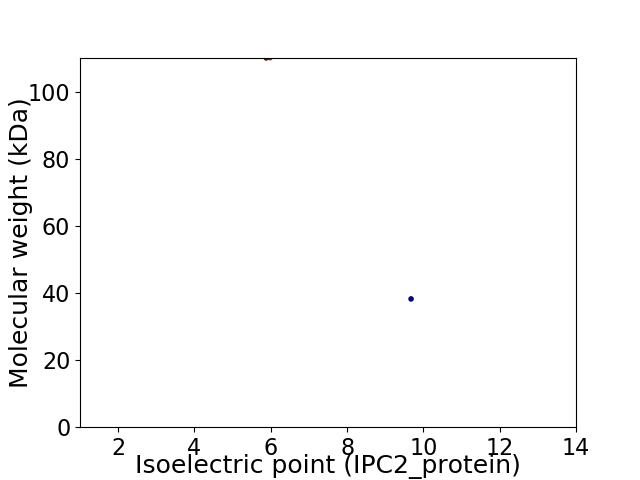
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
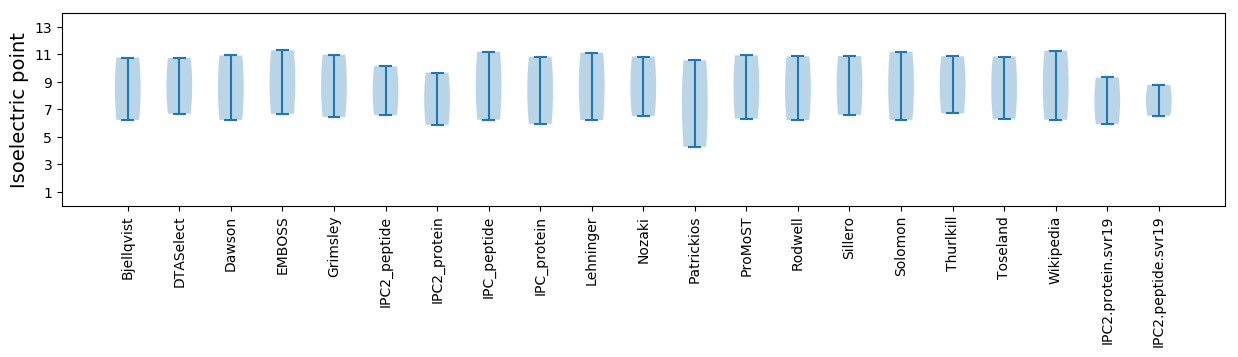
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q3C207|Q3C207_HCRNA RNA-directed RNA polymerase OS=Heterocapsa circularisquama RNA virus 01 (strain 34) OX=1289474 GN=HcRNAV34 ORF-1 PE=4 SV=1
MM1 pKa = 7.28VFLTAAISTSLWATGTTGAVGATTYY26 pKa = 10.67FGVKK30 pKa = 8.58FRR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84VRR36 pKa = 11.84AWLHH40 pKa = 4.28YY41 pKa = 9.38RR42 pKa = 11.84QLNLRR47 pKa = 11.84VGHH50 pKa = 5.57FTSKK54 pKa = 10.74GKK56 pKa = 10.33LFYY59 pKa = 10.85RR60 pKa = 11.84EE61 pKa = 4.4DD62 pKa = 3.95ADD64 pKa = 3.92GPLYY68 pKa = 10.43QAVLKK73 pKa = 10.23RR74 pKa = 11.84SKK76 pKa = 10.66GINPRR81 pKa = 11.84GEE83 pKa = 4.88LIDD86 pKa = 4.09PEE88 pKa = 4.15YY89 pKa = 10.89DD90 pKa = 3.49YY91 pKa = 11.9TEE93 pKa = 4.89LSTSLMSFNEE103 pKa = 3.97SSLVGSTPVRR113 pKa = 11.84RR114 pKa = 11.84IPRR117 pKa = 11.84SLAPSHH123 pKa = 6.34LVVLFSDD130 pKa = 4.1DD131 pKa = 3.46QVSGVGFRR139 pKa = 11.84MGEE142 pKa = 3.81NLITARR148 pKa = 11.84HH149 pKa = 5.34VLDD152 pKa = 3.78LTHH155 pKa = 6.57NLWVQGPRR163 pKa = 11.84GRR165 pKa = 11.84IRR167 pKa = 11.84VEE169 pKa = 4.14EE170 pKa = 4.29PPVTPPVSDD179 pKa = 3.9YY180 pKa = 10.48EE181 pKa = 4.26YY182 pKa = 10.05TGADD186 pKa = 2.6ICYY189 pKa = 9.87IPLTPGQWADD199 pKa = 3.77LGVKK203 pKa = 9.53SMKK206 pKa = 10.25KK207 pKa = 9.76NAVSLSAAGLIKK219 pKa = 10.64LYY221 pKa = 10.87GADD224 pKa = 3.6EE225 pKa = 5.05DD226 pKa = 4.52GLYY229 pKa = 10.86EE230 pKa = 4.75SIGPLKK236 pKa = 10.46VDD238 pKa = 3.81PSLTKK243 pKa = 10.59GKK245 pKa = 10.74GLISYY250 pKa = 7.61VASTTPGFSGSPVLLRR266 pKa = 11.84NAGGGEE272 pKa = 4.48SVVGMHH278 pKa = 6.19VCGDD282 pKa = 3.93FKK284 pKa = 11.59GNGANHH290 pKa = 6.41GVGASSINLILKK302 pKa = 7.82GTGAVPDD309 pKa = 4.46ALTVSDD315 pKa = 4.89FLGPSKK321 pKa = 10.81LEE323 pKa = 3.62SAMYY327 pKa = 10.1YY328 pKa = 10.47DD329 pKa = 3.78KK330 pKa = 11.52QEE332 pKa = 4.51TYY334 pKa = 9.89WDD336 pKa = 3.25WVFDD340 pKa = 3.85QEE342 pKa = 5.26CEE344 pKa = 4.03RR345 pKa = 11.84EE346 pKa = 4.0EE347 pKa = 4.0AARR350 pKa = 11.84EE351 pKa = 4.05AEE353 pKa = 4.12EE354 pKa = 5.33AEE356 pKa = 4.19IEE358 pKa = 4.22HH359 pKa = 7.04LRR361 pKa = 11.84DD362 pKa = 3.3LYY364 pKa = 11.4YY365 pKa = 11.26GEE367 pKa = 4.37TTLALAPVAPPVPSFNGPGARR388 pKa = 11.84ALRR391 pKa = 11.84SYY393 pKa = 11.21NEE395 pKa = 3.92SALNVPARR403 pKa = 11.84LGEE406 pKa = 3.89LSPRR410 pKa = 11.84VGPVEE415 pKa = 4.35SSTHH419 pKa = 5.53LPRR422 pKa = 11.84FLMGASTEE430 pKa = 4.43RR431 pKa = 11.84FWSRR435 pKa = 11.84SINDD439 pKa = 3.81TPPAPLQRR447 pKa = 11.84EE448 pKa = 4.43SPTLKK453 pKa = 9.72ATPEE457 pKa = 4.15LVVSMFTKK465 pKa = 10.23IAEE468 pKa = 4.49GDD470 pKa = 3.45HH471 pKa = 6.55KK472 pKa = 11.36AFLLAIDD479 pKa = 4.28MSYY482 pKa = 11.73DD483 pKa = 3.9DD484 pKa = 5.95LLNCTKK490 pKa = 10.92DD491 pKa = 3.32EE492 pKa = 4.16VMDD495 pKa = 4.37PFSDD499 pKa = 3.53FRR501 pKa = 11.84QYY503 pKa = 11.36LEE505 pKa = 3.98HH506 pKa = 6.33TRR508 pKa = 11.84KK509 pKa = 9.88LMKK512 pKa = 10.65KK513 pKa = 9.12NDD515 pKa = 3.74GGRR518 pKa = 11.84EE519 pKa = 3.87LSDD522 pKa = 3.07QSGRR526 pKa = 11.84TFFRR530 pKa = 11.84EE531 pKa = 3.94THH533 pKa = 5.92RR534 pKa = 11.84TTQSGGGKK542 pKa = 9.59KK543 pKa = 8.32RR544 pKa = 11.84TAPVWLNEE552 pKa = 3.78EE553 pKa = 3.98SKK555 pKa = 11.34AFLRR559 pKa = 11.84GLGVEE564 pKa = 4.67GEE566 pKa = 4.47YY567 pKa = 10.71CRR569 pKa = 11.84PPNDD573 pKa = 3.31EE574 pKa = 3.91ASIIRR579 pKa = 11.84SMEE582 pKa = 3.69NQAARR587 pKa = 11.84QLGSRR592 pKa = 11.84AWPSTGAMEE601 pKa = 4.6PFLVQYY607 pKa = 10.72LGDD610 pKa = 3.99AAVSPPTFSPGRR622 pKa = 11.84QGFDD626 pKa = 2.83QLYY629 pKa = 10.2RR630 pKa = 11.84SFDD633 pKa = 3.6DD634 pKa = 5.67SSAGWTRR641 pKa = 11.84RR642 pKa = 11.84YY643 pKa = 10.52RR644 pKa = 11.84NLTKK648 pKa = 10.2KK649 pKa = 10.42SYY651 pKa = 11.09VSGPHH656 pKa = 6.29CGEE659 pKa = 3.83LTRR662 pKa = 11.84IAVARR667 pKa = 11.84ILLRR671 pKa = 11.84CTMWHH676 pKa = 7.51RR677 pKa = 11.84IPSMTPEE684 pKa = 3.81EE685 pKa = 4.41MVWLGLRR692 pKa = 11.84DD693 pKa = 3.72PTDD696 pKa = 3.17VFVKK700 pKa = 10.57DD701 pKa = 4.17EE702 pKa = 4.03PHH704 pKa = 6.1TADD707 pKa = 3.35KK708 pKa = 10.86ARR710 pKa = 11.84RR711 pKa = 11.84EE712 pKa = 3.91MWRR715 pKa = 11.84LIWNVSLVDD724 pKa = 4.8SICQAYY730 pKa = 9.65FNRR733 pKa = 11.84EE734 pKa = 3.51LNLQQNRR741 pKa = 11.84DD742 pKa = 3.79YY743 pKa = 11.29QGGHH747 pKa = 6.53PVPHH751 pKa = 5.99TCGMGHH757 pKa = 6.82HH758 pKa = 7.44DD759 pKa = 4.27EE760 pKa = 4.96GIKK763 pKa = 10.55RR764 pKa = 11.84LGEE767 pKa = 4.31AIEE770 pKa = 4.04AAFPDD775 pKa = 5.22GIVCSSDD782 pKa = 2.87ASGWDD787 pKa = 3.24MSVSRR792 pKa = 11.84DD793 pKa = 3.27GLIFDD798 pKa = 4.06GLVRR802 pKa = 11.84AIRR805 pKa = 11.84TQDD808 pKa = 3.33PSATAYY814 pKa = 10.58HH815 pKa = 6.58NIGTSVCILLDD826 pKa = 3.43KK827 pKa = 10.45FVQSAHH833 pKa = 6.22MICTGTSLWSVDD845 pKa = 3.44VYY847 pKa = 11.4GITASGLPDD856 pKa = 3.25TTTQNSFVRR865 pKa = 11.84GMGAKK870 pKa = 10.06LAGCFKK876 pKa = 10.97ALTAGDD882 pKa = 5.61DD883 pKa = 3.92LLCDD887 pKa = 3.33NRR889 pKa = 11.84LRR891 pKa = 11.84LPVLTEE897 pKa = 3.97HH898 pKa = 6.15GTITKK903 pKa = 10.69GDD905 pKa = 3.49VTTANWRR912 pKa = 11.84KK913 pKa = 8.04GQPVGFTSHH922 pKa = 6.3SLVRR926 pKa = 11.84GPDD929 pKa = 4.08GQWSAHH935 pKa = 5.74FEE937 pKa = 4.23NEE939 pKa = 3.79SKK941 pKa = 10.84ALHH944 pKa = 6.33RR945 pKa = 11.84LLLSGKK951 pKa = 8.0TPLPEE956 pKa = 4.1QIGGVAFAEE965 pKa = 4.44RR966 pKa = 11.84KK967 pKa = 10.19DD968 pKa = 3.7EE969 pKa = 4.32AQLGRR974 pKa = 11.84LRR976 pKa = 11.84AVCDD980 pKa = 3.5SRR982 pKa = 11.84GWPVPPQSEE991 pKa = 4.06WAEE994 pKa = 4.2DD995 pKa = 3.56FDD997 pKa = 5.27LCC999 pKa = 5.79
MM1 pKa = 7.28VFLTAAISTSLWATGTTGAVGATTYY26 pKa = 10.67FGVKK30 pKa = 8.58FRR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84VRR36 pKa = 11.84AWLHH40 pKa = 4.28YY41 pKa = 9.38RR42 pKa = 11.84QLNLRR47 pKa = 11.84VGHH50 pKa = 5.57FTSKK54 pKa = 10.74GKK56 pKa = 10.33LFYY59 pKa = 10.85RR60 pKa = 11.84EE61 pKa = 4.4DD62 pKa = 3.95ADD64 pKa = 3.92GPLYY68 pKa = 10.43QAVLKK73 pKa = 10.23RR74 pKa = 11.84SKK76 pKa = 10.66GINPRR81 pKa = 11.84GEE83 pKa = 4.88LIDD86 pKa = 4.09PEE88 pKa = 4.15YY89 pKa = 10.89DD90 pKa = 3.49YY91 pKa = 11.9TEE93 pKa = 4.89LSTSLMSFNEE103 pKa = 3.97SSLVGSTPVRR113 pKa = 11.84RR114 pKa = 11.84IPRR117 pKa = 11.84SLAPSHH123 pKa = 6.34LVVLFSDD130 pKa = 4.1DD131 pKa = 3.46QVSGVGFRR139 pKa = 11.84MGEE142 pKa = 3.81NLITARR148 pKa = 11.84HH149 pKa = 5.34VLDD152 pKa = 3.78LTHH155 pKa = 6.57NLWVQGPRR163 pKa = 11.84GRR165 pKa = 11.84IRR167 pKa = 11.84VEE169 pKa = 4.14EE170 pKa = 4.29PPVTPPVSDD179 pKa = 3.9YY180 pKa = 10.48EE181 pKa = 4.26YY182 pKa = 10.05TGADD186 pKa = 2.6ICYY189 pKa = 9.87IPLTPGQWADD199 pKa = 3.77LGVKK203 pKa = 9.53SMKK206 pKa = 10.25KK207 pKa = 9.76NAVSLSAAGLIKK219 pKa = 10.64LYY221 pKa = 10.87GADD224 pKa = 3.6EE225 pKa = 5.05DD226 pKa = 4.52GLYY229 pKa = 10.86EE230 pKa = 4.75SIGPLKK236 pKa = 10.46VDD238 pKa = 3.81PSLTKK243 pKa = 10.59GKK245 pKa = 10.74GLISYY250 pKa = 7.61VASTTPGFSGSPVLLRR266 pKa = 11.84NAGGGEE272 pKa = 4.48SVVGMHH278 pKa = 6.19VCGDD282 pKa = 3.93FKK284 pKa = 11.59GNGANHH290 pKa = 6.41GVGASSINLILKK302 pKa = 7.82GTGAVPDD309 pKa = 4.46ALTVSDD315 pKa = 4.89FLGPSKK321 pKa = 10.81LEE323 pKa = 3.62SAMYY327 pKa = 10.1YY328 pKa = 10.47DD329 pKa = 3.78KK330 pKa = 11.52QEE332 pKa = 4.51TYY334 pKa = 9.89WDD336 pKa = 3.25WVFDD340 pKa = 3.85QEE342 pKa = 5.26CEE344 pKa = 4.03RR345 pKa = 11.84EE346 pKa = 4.0EE347 pKa = 4.0AARR350 pKa = 11.84EE351 pKa = 4.05AEE353 pKa = 4.12EE354 pKa = 5.33AEE356 pKa = 4.19IEE358 pKa = 4.22HH359 pKa = 7.04LRR361 pKa = 11.84DD362 pKa = 3.3LYY364 pKa = 11.4YY365 pKa = 11.26GEE367 pKa = 4.37TTLALAPVAPPVPSFNGPGARR388 pKa = 11.84ALRR391 pKa = 11.84SYY393 pKa = 11.21NEE395 pKa = 3.92SALNVPARR403 pKa = 11.84LGEE406 pKa = 3.89LSPRR410 pKa = 11.84VGPVEE415 pKa = 4.35SSTHH419 pKa = 5.53LPRR422 pKa = 11.84FLMGASTEE430 pKa = 4.43RR431 pKa = 11.84FWSRR435 pKa = 11.84SINDD439 pKa = 3.81TPPAPLQRR447 pKa = 11.84EE448 pKa = 4.43SPTLKK453 pKa = 9.72ATPEE457 pKa = 4.15LVVSMFTKK465 pKa = 10.23IAEE468 pKa = 4.49GDD470 pKa = 3.45HH471 pKa = 6.55KK472 pKa = 11.36AFLLAIDD479 pKa = 4.28MSYY482 pKa = 11.73DD483 pKa = 3.9DD484 pKa = 5.95LLNCTKK490 pKa = 10.92DD491 pKa = 3.32EE492 pKa = 4.16VMDD495 pKa = 4.37PFSDD499 pKa = 3.53FRR501 pKa = 11.84QYY503 pKa = 11.36LEE505 pKa = 3.98HH506 pKa = 6.33TRR508 pKa = 11.84KK509 pKa = 9.88LMKK512 pKa = 10.65KK513 pKa = 9.12NDD515 pKa = 3.74GGRR518 pKa = 11.84EE519 pKa = 3.87LSDD522 pKa = 3.07QSGRR526 pKa = 11.84TFFRR530 pKa = 11.84EE531 pKa = 3.94THH533 pKa = 5.92RR534 pKa = 11.84TTQSGGGKK542 pKa = 9.59KK543 pKa = 8.32RR544 pKa = 11.84TAPVWLNEE552 pKa = 3.78EE553 pKa = 3.98SKK555 pKa = 11.34AFLRR559 pKa = 11.84GLGVEE564 pKa = 4.67GEE566 pKa = 4.47YY567 pKa = 10.71CRR569 pKa = 11.84PPNDD573 pKa = 3.31EE574 pKa = 3.91ASIIRR579 pKa = 11.84SMEE582 pKa = 3.69NQAARR587 pKa = 11.84QLGSRR592 pKa = 11.84AWPSTGAMEE601 pKa = 4.6PFLVQYY607 pKa = 10.72LGDD610 pKa = 3.99AAVSPPTFSPGRR622 pKa = 11.84QGFDD626 pKa = 2.83QLYY629 pKa = 10.2RR630 pKa = 11.84SFDD633 pKa = 3.6DD634 pKa = 5.67SSAGWTRR641 pKa = 11.84RR642 pKa = 11.84YY643 pKa = 10.52RR644 pKa = 11.84NLTKK648 pKa = 10.2KK649 pKa = 10.42SYY651 pKa = 11.09VSGPHH656 pKa = 6.29CGEE659 pKa = 3.83LTRR662 pKa = 11.84IAVARR667 pKa = 11.84ILLRR671 pKa = 11.84CTMWHH676 pKa = 7.51RR677 pKa = 11.84IPSMTPEE684 pKa = 3.81EE685 pKa = 4.41MVWLGLRR692 pKa = 11.84DD693 pKa = 3.72PTDD696 pKa = 3.17VFVKK700 pKa = 10.57DD701 pKa = 4.17EE702 pKa = 4.03PHH704 pKa = 6.1TADD707 pKa = 3.35KK708 pKa = 10.86ARR710 pKa = 11.84RR711 pKa = 11.84EE712 pKa = 3.91MWRR715 pKa = 11.84LIWNVSLVDD724 pKa = 4.8SICQAYY730 pKa = 9.65FNRR733 pKa = 11.84EE734 pKa = 3.51LNLQQNRR741 pKa = 11.84DD742 pKa = 3.79YY743 pKa = 11.29QGGHH747 pKa = 6.53PVPHH751 pKa = 5.99TCGMGHH757 pKa = 6.82HH758 pKa = 7.44DD759 pKa = 4.27EE760 pKa = 4.96GIKK763 pKa = 10.55RR764 pKa = 11.84LGEE767 pKa = 4.31AIEE770 pKa = 4.04AAFPDD775 pKa = 5.22GIVCSSDD782 pKa = 2.87ASGWDD787 pKa = 3.24MSVSRR792 pKa = 11.84DD793 pKa = 3.27GLIFDD798 pKa = 4.06GLVRR802 pKa = 11.84AIRR805 pKa = 11.84TQDD808 pKa = 3.33PSATAYY814 pKa = 10.58HH815 pKa = 6.58NIGTSVCILLDD826 pKa = 3.43KK827 pKa = 10.45FVQSAHH833 pKa = 6.22MICTGTSLWSVDD845 pKa = 3.44VYY847 pKa = 11.4GITASGLPDD856 pKa = 3.25TTTQNSFVRR865 pKa = 11.84GMGAKK870 pKa = 10.06LAGCFKK876 pKa = 10.97ALTAGDD882 pKa = 5.61DD883 pKa = 3.92LLCDD887 pKa = 3.33NRR889 pKa = 11.84LRR891 pKa = 11.84LPVLTEE897 pKa = 3.97HH898 pKa = 6.15GTITKK903 pKa = 10.69GDD905 pKa = 3.49VTTANWRR912 pKa = 11.84KK913 pKa = 8.04GQPVGFTSHH922 pKa = 6.3SLVRR926 pKa = 11.84GPDD929 pKa = 4.08GQWSAHH935 pKa = 5.74FEE937 pKa = 4.23NEE939 pKa = 3.79SKK941 pKa = 10.84ALHH944 pKa = 6.33RR945 pKa = 11.84LLLSGKK951 pKa = 8.0TPLPEE956 pKa = 4.1QIGGVAFAEE965 pKa = 4.44RR966 pKa = 11.84KK967 pKa = 10.19DD968 pKa = 3.7EE969 pKa = 4.32AQLGRR974 pKa = 11.84LRR976 pKa = 11.84AVCDD980 pKa = 3.5SRR982 pKa = 11.84GWPVPPQSEE991 pKa = 4.06WAEE994 pKa = 4.2DD995 pKa = 3.56FDD997 pKa = 5.27LCC999 pKa = 5.79
Molecular weight: 110.08 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q3C207|Q3C207_HCRNA RNA-directed RNA polymerase OS=Heterocapsa circularisquama RNA virus 01 (strain 34) OX=1289474 GN=HcRNAV34 ORF-1 PE=4 SV=1
MM1 pKa = 7.25TRR3 pKa = 11.84PLALTNGGNTNGGNNGGSRR22 pKa = 11.84RR23 pKa = 11.84RR24 pKa = 11.84PPRR27 pKa = 11.84QRR29 pKa = 11.84RR30 pKa = 11.84QGRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84NRR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84GGGGGGPRR47 pKa = 11.84NNAAMVLAQGAGSVPGMPFGSWPSRR72 pKa = 11.84STMRR76 pKa = 11.84AWDD79 pKa = 3.7AFHH82 pKa = 7.53PEE84 pKa = 4.35HH85 pKa = 6.81LPLPRR90 pKa = 11.84SVGPYY95 pKa = 9.14CVVRR99 pKa = 11.84TSSLITSSDD108 pKa = 2.93KK109 pKa = 11.5VMLFAPMVGSAGCWLTACAMGSRR132 pKa = 11.84TEE134 pKa = 4.05GGAINGLDD142 pKa = 3.45NTNVYY147 pKa = 8.22TVPFPGIATTGSSITVVPAALSVQVMNPNPLMSTTGIFGGTVSHH191 pKa = 6.25TQLNLAGRR199 pKa = 11.84TEE201 pKa = 3.96TWNDD205 pKa = 2.75FSMEE209 pKa = 4.36VISFMRR215 pKa = 11.84PRR217 pKa = 11.84LMSAGKK223 pKa = 9.4LALRR227 pKa = 11.84GVQGDD232 pKa = 4.24SYY234 pKa = 9.57PLNMSALSNFNCLSDD249 pKa = 3.7VAEE252 pKa = 5.38GKK254 pKa = 10.69LSWTDD259 pKa = 3.28SSGHH263 pKa = 5.54YY264 pKa = 9.38PAGLAPLVFVNEE276 pKa = 4.63AKK278 pKa = 8.88QTMNYY283 pKa = 9.07LVSVEE288 pKa = 3.55WRR290 pKa = 11.84VRR292 pKa = 11.84FDD294 pKa = 2.93IGNPAVAAQRR304 pKa = 11.84HH305 pKa = 5.57HH306 pKa = 7.3GITPEE311 pKa = 4.07WKK313 pKa = 8.29WDD315 pKa = 3.79DD316 pKa = 3.88MIKK319 pKa = 9.12TAIARR324 pKa = 11.84GHH326 pKa = 6.83GIMDD330 pKa = 3.46IAEE333 pKa = 4.04RR334 pKa = 11.84VANAGSFVANAAIAARR350 pKa = 11.84RR351 pKa = 11.84AMPALMAAA359 pKa = 4.3
MM1 pKa = 7.25TRR3 pKa = 11.84PLALTNGGNTNGGNNGGSRR22 pKa = 11.84RR23 pKa = 11.84RR24 pKa = 11.84PPRR27 pKa = 11.84QRR29 pKa = 11.84RR30 pKa = 11.84QGRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84NRR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84GGGGGGPRR47 pKa = 11.84NNAAMVLAQGAGSVPGMPFGSWPSRR72 pKa = 11.84STMRR76 pKa = 11.84AWDD79 pKa = 3.7AFHH82 pKa = 7.53PEE84 pKa = 4.35HH85 pKa = 6.81LPLPRR90 pKa = 11.84SVGPYY95 pKa = 9.14CVVRR99 pKa = 11.84TSSLITSSDD108 pKa = 2.93KK109 pKa = 11.5VMLFAPMVGSAGCWLTACAMGSRR132 pKa = 11.84TEE134 pKa = 4.05GGAINGLDD142 pKa = 3.45NTNVYY147 pKa = 8.22TVPFPGIATTGSSITVVPAALSVQVMNPNPLMSTTGIFGGTVSHH191 pKa = 6.25TQLNLAGRR199 pKa = 11.84TEE201 pKa = 3.96TWNDD205 pKa = 2.75FSMEE209 pKa = 4.36VISFMRR215 pKa = 11.84PRR217 pKa = 11.84LMSAGKK223 pKa = 9.4LALRR227 pKa = 11.84GVQGDD232 pKa = 4.24SYY234 pKa = 9.57PLNMSALSNFNCLSDD249 pKa = 3.7VAEE252 pKa = 5.38GKK254 pKa = 10.69LSWTDD259 pKa = 3.28SSGHH263 pKa = 5.54YY264 pKa = 9.38PAGLAPLVFVNEE276 pKa = 4.63AKK278 pKa = 8.88QTMNYY283 pKa = 9.07LVSVEE288 pKa = 3.55WRR290 pKa = 11.84VRR292 pKa = 11.84FDD294 pKa = 2.93IGNPAVAAQRR304 pKa = 11.84HH305 pKa = 5.57HH306 pKa = 7.3GITPEE311 pKa = 4.07WKK313 pKa = 8.29WDD315 pKa = 3.79DD316 pKa = 3.88MIKK319 pKa = 9.12TAIARR324 pKa = 11.84GHH326 pKa = 6.83GIMDD330 pKa = 3.46IAEE333 pKa = 4.04RR334 pKa = 11.84VANAGSFVANAAIAARR350 pKa = 11.84RR351 pKa = 11.84AMPALMAAA359 pKa = 4.3
Molecular weight: 38.24 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1358 |
359 |
999 |
679.0 |
74.16 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.542 ± 1.373 | 1.473 ± 0.189 |
5.228 ± 1.143 | 5.007 ± 1.32 |
3.535 ± 0.248 | 9.794 ± 0.859 |
2.356 ± 0.215 | 3.535 ± 0.046 |
3.314 ± 0.867 | 8.763 ± 1.097 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.872 ± 1.131 | 3.829 ± 1.361 |
6.186 ± 0.264 | 2.651 ± 0.223 |
7.364 ± 0.524 | 8.027 ± 0.027 |
6.406 ± 0.0 | 6.554 ± 0.216 |
2.062 ± 0.088 | 2.504 ± 0.587 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
