
Clostridium sp. CAG:58
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Clostridiaceae; Clostridium; environmental samples
Average proteome isoelectric point is 6.1
Get precalculated fractions of proteins
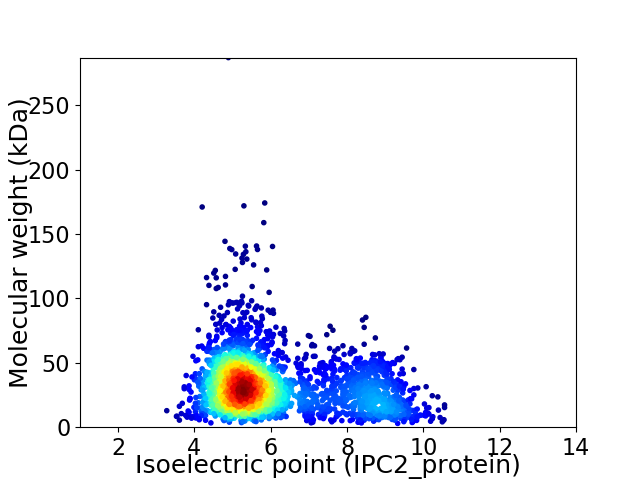
Virtual 2D-PAGE plot for 2660 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
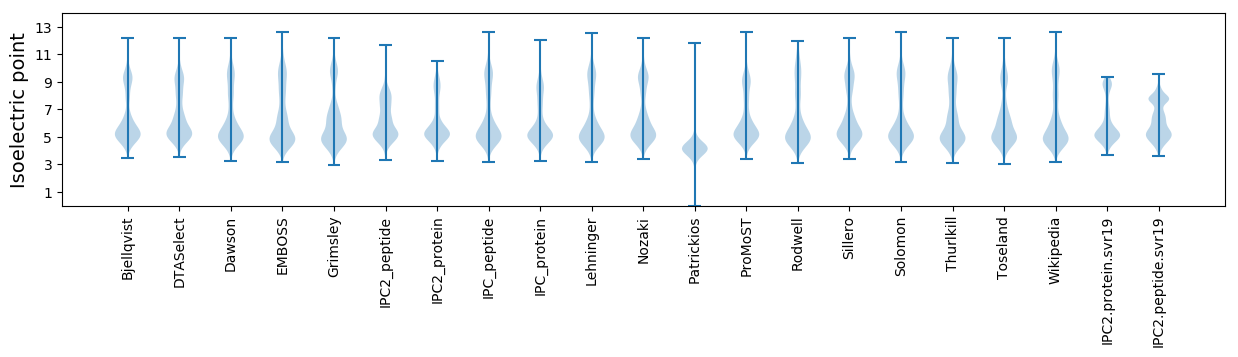
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R6RLV8|R6RLV8_9CLOT Branched-chain amino acid transport system carrier protein OS=Clostridium sp. CAG:58 OX=1262824 GN=BN719_02356 PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 10.17KK3 pKa = 10.45KK4 pKa = 10.63IMTLVLTTAMTAAAGLTAFAGEE26 pKa = 4.34WKK28 pKa = 10.9SNDD31 pKa = 3.67TGWWWLNDD39 pKa = 3.87DD40 pKa = 4.54GSNPAAEE47 pKa = 4.48WKK49 pKa = 9.65WLDD52 pKa = 3.5GNKK55 pKa = 10.49DD56 pKa = 3.56GTFEE60 pKa = 4.35CYY62 pKa = 10.54YY63 pKa = 10.2FGNDD67 pKa = 3.66GYY69 pKa = 10.3MLSDD73 pKa = 3.05TTTPDD78 pKa = 2.83NYY80 pKa = 10.53QVNKK84 pKa = 10.2DD85 pKa = 3.67GAWIVDD91 pKa = 3.58GGVQLQFNLEE101 pKa = 4.01EE102 pKa = 4.02NAGNDD107 pKa = 3.62EE108 pKa = 4.16EE109 pKa = 6.16DD110 pKa = 4.12GEE112 pKa = 4.53NSGEE116 pKa = 3.73ASYY119 pKa = 10.96EE120 pKa = 3.64YY121 pKa = 10.43DD122 pKa = 3.59AYY124 pKa = 11.14DD125 pKa = 5.11LIGTYY130 pKa = 8.39TSEE133 pKa = 4.93DD134 pKa = 3.38GDD136 pKa = 3.96YY137 pKa = 10.91MEE139 pKa = 6.19LNLGADD145 pKa = 3.77DD146 pKa = 4.75SLVGMYY152 pKa = 10.19YY153 pKa = 10.56DD154 pKa = 4.38EE155 pKa = 6.18DD156 pKa = 4.53GSFDD160 pKa = 4.1GIYY163 pKa = 10.43SFKK166 pKa = 10.61KK167 pKa = 10.46VNSLEE172 pKa = 4.34YY173 pKa = 10.22KK174 pKa = 10.56DD175 pKa = 4.78SLHH178 pKa = 6.27SRR180 pKa = 11.84SIKK183 pKa = 10.15FSNEE187 pKa = 2.91GSFTMRR193 pKa = 11.84DD194 pKa = 3.06TVYY197 pKa = 10.88YY198 pKa = 10.3RR199 pKa = 3.59
MM1 pKa = 7.45KK2 pKa = 10.17KK3 pKa = 10.45KK4 pKa = 10.63IMTLVLTTAMTAAAGLTAFAGEE26 pKa = 4.34WKK28 pKa = 10.9SNDD31 pKa = 3.67TGWWWLNDD39 pKa = 3.87DD40 pKa = 4.54GSNPAAEE47 pKa = 4.48WKK49 pKa = 9.65WLDD52 pKa = 3.5GNKK55 pKa = 10.49DD56 pKa = 3.56GTFEE60 pKa = 4.35CYY62 pKa = 10.54YY63 pKa = 10.2FGNDD67 pKa = 3.66GYY69 pKa = 10.3MLSDD73 pKa = 3.05TTTPDD78 pKa = 2.83NYY80 pKa = 10.53QVNKK84 pKa = 10.2DD85 pKa = 3.67GAWIVDD91 pKa = 3.58GGVQLQFNLEE101 pKa = 4.01EE102 pKa = 4.02NAGNDD107 pKa = 3.62EE108 pKa = 4.16EE109 pKa = 6.16DD110 pKa = 4.12GEE112 pKa = 4.53NSGEE116 pKa = 3.73ASYY119 pKa = 10.96EE120 pKa = 3.64YY121 pKa = 10.43DD122 pKa = 3.59AYY124 pKa = 11.14DD125 pKa = 5.11LIGTYY130 pKa = 8.39TSEE133 pKa = 4.93DD134 pKa = 3.38GDD136 pKa = 3.96YY137 pKa = 10.91MEE139 pKa = 6.19LNLGADD145 pKa = 3.77DD146 pKa = 4.75SLVGMYY152 pKa = 10.19YY153 pKa = 10.56DD154 pKa = 4.38EE155 pKa = 6.18DD156 pKa = 4.53GSFDD160 pKa = 4.1GIYY163 pKa = 10.43SFKK166 pKa = 10.61KK167 pKa = 10.46VNSLEE172 pKa = 4.34YY173 pKa = 10.22KK174 pKa = 10.56DD175 pKa = 4.78SLHH178 pKa = 6.27SRR180 pKa = 11.84SIKK183 pKa = 10.15FSNEE187 pKa = 2.91GSFTMRR193 pKa = 11.84DD194 pKa = 3.06TVYY197 pKa = 10.88YY198 pKa = 10.3RR199 pKa = 3.59
Molecular weight: 22.43 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R6S4K1|R6S4K1_9CLOT SpoIID/LytB domain OS=Clostridium sp. CAG:58 OX=1262824 GN=BN719_00370 PE=4 SV=1
MM1 pKa = 7.5GGHH4 pKa = 6.53RR5 pKa = 11.84LSRR8 pKa = 11.84HH9 pKa = 4.56LHH11 pKa = 5.89IDD13 pKa = 3.08PLVIRR18 pKa = 11.84RR19 pKa = 11.84HH20 pKa = 5.25RR21 pKa = 11.84RR22 pKa = 11.84IQGRR26 pKa = 11.84RR27 pKa = 11.84HH28 pKa = 4.84SRR30 pKa = 11.84IVDD33 pKa = 3.95RR34 pKa = 11.84FRR36 pKa = 11.84SLSQRR41 pKa = 11.84VHH43 pKa = 6.84AGQINVILCLILQPLKK59 pKa = 10.57LIPVLPDD66 pKa = 2.82IRR68 pKa = 11.84DD69 pKa = 3.42LRR71 pKa = 11.84VRR73 pKa = 11.84SLGVAVLPVIQVIAVCIHH91 pKa = 6.9HH92 pKa = 7.19ILPADD97 pKa = 4.54GKK99 pKa = 9.39PLIPGCHH106 pKa = 6.14RR107 pKa = 11.84DD108 pKa = 2.85IFRR111 pKa = 11.84CRR113 pKa = 11.84RR114 pKa = 11.84KK115 pKa = 10.03AGRR118 pKa = 11.84PGLSHH123 pKa = 6.08SLCRR127 pKa = 11.84RR128 pKa = 11.84ALPGPVHH135 pKa = 6.38RR136 pKa = 11.84TDD138 pKa = 4.32LDD140 pKa = 4.45LIGISIGQRR149 pKa = 11.84PVSQAFCPEE158 pKa = 3.34FRR160 pKa = 11.84LIRR163 pKa = 11.84LEE165 pKa = 4.09GLCHH169 pKa = 7.22LVFLTVDD176 pKa = 2.8SMDD179 pKa = 3.08RR180 pKa = 11.84HH181 pKa = 5.49LVARR185 pKa = 11.84HH186 pKa = 6.14IGLSPPGHH194 pKa = 5.86GKK196 pKa = 10.1GRR198 pKa = 11.84VPVLHH203 pKa = 6.45LHH205 pKa = 6.06PLRR208 pKa = 11.84RR209 pKa = 11.84CHH211 pKa = 6.82FRR213 pKa = 11.84LRR215 pKa = 11.84DD216 pKa = 3.7PFLHH220 pKa = 6.4VPQEE224 pKa = 3.81LHH226 pKa = 6.53LCNIHH231 pKa = 6.41QGSVPQGPADD241 pKa = 3.92LNGHH245 pKa = 6.55LLHH248 pKa = 7.04GCLALQIDD256 pKa = 4.77DD257 pKa = 5.41DD258 pKa = 4.71GASLAYY264 pKa = 10.06GLLFSCEE271 pKa = 4.07KK272 pKa = 9.48FQKK275 pKa = 10.29RR276 pKa = 11.84PSQAPFLYY284 pKa = 10.4FSVFDD289 pKa = 4.41LKK291 pKa = 11.42DD292 pKa = 3.28LFFSLFCLHH301 pKa = 6.82RR302 pKa = 11.84NSSVHH307 pKa = 5.98NGVARR312 pKa = 11.84AVHH315 pKa = 6.23RR316 pKa = 11.84VDD318 pKa = 5.49DD319 pKa = 4.47EE320 pKa = 4.82PGHH323 pKa = 5.89GLSGAKK329 pKa = 9.59LRR331 pKa = 11.84RR332 pKa = 11.84NGHH335 pKa = 6.7RR336 pKa = 11.84ILPGNGLLTFQGKK349 pKa = 9.86AGRR352 pKa = 11.84TGKK355 pKa = 9.84QLLHH359 pKa = 6.95RR360 pKa = 11.84SLGDD364 pKa = 3.28LRR366 pKa = 11.84VAGGRR371 pKa = 11.84RR372 pKa = 11.84MDD374 pKa = 5.46LFLCDD379 pKa = 3.48PHH381 pKa = 8.78GPDD384 pKa = 5.1LPGSPYY390 pKa = 9.97IFLIILCKK398 pKa = 8.94TCYY401 pKa = 9.76VRR403 pKa = 11.84QIHH406 pKa = 5.83LTVAVQIGPVIKK418 pKa = 10.43QIVMDD423 pKa = 4.67LAGGCQLQKK432 pKa = 11.18LLLIVLIHH440 pKa = 6.6SSAAVDD446 pKa = 3.3IAHH449 pKa = 6.49GQNADD454 pKa = 3.56AASGSALPVSVVFRR468 pKa = 11.84RR469 pKa = 11.84IVHH472 pKa = 6.26
MM1 pKa = 7.5GGHH4 pKa = 6.53RR5 pKa = 11.84LSRR8 pKa = 11.84HH9 pKa = 4.56LHH11 pKa = 5.89IDD13 pKa = 3.08PLVIRR18 pKa = 11.84RR19 pKa = 11.84HH20 pKa = 5.25RR21 pKa = 11.84RR22 pKa = 11.84IQGRR26 pKa = 11.84RR27 pKa = 11.84HH28 pKa = 4.84SRR30 pKa = 11.84IVDD33 pKa = 3.95RR34 pKa = 11.84FRR36 pKa = 11.84SLSQRR41 pKa = 11.84VHH43 pKa = 6.84AGQINVILCLILQPLKK59 pKa = 10.57LIPVLPDD66 pKa = 2.82IRR68 pKa = 11.84DD69 pKa = 3.42LRR71 pKa = 11.84VRR73 pKa = 11.84SLGVAVLPVIQVIAVCIHH91 pKa = 6.9HH92 pKa = 7.19ILPADD97 pKa = 4.54GKK99 pKa = 9.39PLIPGCHH106 pKa = 6.14RR107 pKa = 11.84DD108 pKa = 2.85IFRR111 pKa = 11.84CRR113 pKa = 11.84RR114 pKa = 11.84KK115 pKa = 10.03AGRR118 pKa = 11.84PGLSHH123 pKa = 6.08SLCRR127 pKa = 11.84RR128 pKa = 11.84ALPGPVHH135 pKa = 6.38RR136 pKa = 11.84TDD138 pKa = 4.32LDD140 pKa = 4.45LIGISIGQRR149 pKa = 11.84PVSQAFCPEE158 pKa = 3.34FRR160 pKa = 11.84LIRR163 pKa = 11.84LEE165 pKa = 4.09GLCHH169 pKa = 7.22LVFLTVDD176 pKa = 2.8SMDD179 pKa = 3.08RR180 pKa = 11.84HH181 pKa = 5.49LVARR185 pKa = 11.84HH186 pKa = 6.14IGLSPPGHH194 pKa = 5.86GKK196 pKa = 10.1GRR198 pKa = 11.84VPVLHH203 pKa = 6.45LHH205 pKa = 6.06PLRR208 pKa = 11.84RR209 pKa = 11.84CHH211 pKa = 6.82FRR213 pKa = 11.84LRR215 pKa = 11.84DD216 pKa = 3.7PFLHH220 pKa = 6.4VPQEE224 pKa = 3.81LHH226 pKa = 6.53LCNIHH231 pKa = 6.41QGSVPQGPADD241 pKa = 3.92LNGHH245 pKa = 6.55LLHH248 pKa = 7.04GCLALQIDD256 pKa = 4.77DD257 pKa = 5.41DD258 pKa = 4.71GASLAYY264 pKa = 10.06GLLFSCEE271 pKa = 4.07KK272 pKa = 9.48FQKK275 pKa = 10.29RR276 pKa = 11.84PSQAPFLYY284 pKa = 10.4FSVFDD289 pKa = 4.41LKK291 pKa = 11.42DD292 pKa = 3.28LFFSLFCLHH301 pKa = 6.82RR302 pKa = 11.84NSSVHH307 pKa = 5.98NGVARR312 pKa = 11.84AVHH315 pKa = 6.23RR316 pKa = 11.84VDD318 pKa = 5.49DD319 pKa = 4.47EE320 pKa = 4.82PGHH323 pKa = 5.89GLSGAKK329 pKa = 9.59LRR331 pKa = 11.84RR332 pKa = 11.84NGHH335 pKa = 6.7RR336 pKa = 11.84ILPGNGLLTFQGKK349 pKa = 9.86AGRR352 pKa = 11.84TGKK355 pKa = 9.84QLLHH359 pKa = 6.95RR360 pKa = 11.84SLGDD364 pKa = 3.28LRR366 pKa = 11.84VAGGRR371 pKa = 11.84RR372 pKa = 11.84MDD374 pKa = 5.46LFLCDD379 pKa = 3.48PHH381 pKa = 8.78GPDD384 pKa = 5.1LPGSPYY390 pKa = 9.97IFLIILCKK398 pKa = 8.94TCYY401 pKa = 9.76VRR403 pKa = 11.84QIHH406 pKa = 5.83LTVAVQIGPVIKK418 pKa = 10.43QIVMDD423 pKa = 4.67LAGGCQLQKK432 pKa = 11.18LLLIVLIHH440 pKa = 6.6SSAAVDD446 pKa = 3.3IAHH449 pKa = 6.49GQNADD454 pKa = 3.56AASGSALPVSVVFRR468 pKa = 11.84RR469 pKa = 11.84IVHH472 pKa = 6.26
Molecular weight: 52.37 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
805197 |
29 |
2543 |
302.7 |
33.65 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.423 ± 0.053 | 1.477 ± 0.02 |
5.452 ± 0.037 | 7.688 ± 0.063 |
4.084 ± 0.03 | 8.12 ± 0.047 |
1.74 ± 0.019 | 6.373 ± 0.046 |
5.845 ± 0.043 | 9.239 ± 0.056 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.284 ± 0.026 | 3.545 ± 0.031 |
3.805 ± 0.027 | 2.979 ± 0.025 |
5.242 ± 0.039 | 5.694 ± 0.042 |
5.232 ± 0.032 | 7.071 ± 0.041 |
0.97 ± 0.018 | 3.727 ± 0.034 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
