
Sowbane mosaic virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Sobelivirales; Solemoviridae; Sobemovirus
Average proteome isoelectric point is 7.12
Get precalculated fractions of proteins
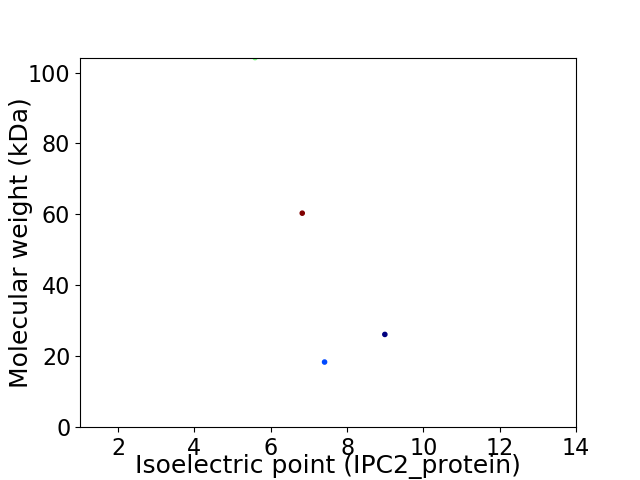
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|B5DC59|B5DC59_9VIRU Capsid protein OS=Sowbane mosaic virus OX=378833 GN=CP PE=3 SV=1
MM1 pKa = 7.15HH2 pKa = 7.85RR3 pKa = 11.84SPFGYY8 pKa = 10.5LLLLISLPAGAVTYY22 pKa = 10.62DD23 pKa = 4.78CIRR26 pKa = 11.84GDD28 pKa = 3.99CQAGPNSLMMALMTLLTTGMWWVVSLSLFWIEE60 pKa = 3.78RR61 pKa = 11.84QRR63 pKa = 11.84YY64 pKa = 8.18RR65 pKa = 11.84SAEE68 pKa = 4.05EE69 pKa = 4.11KK70 pKa = 10.04ISRR73 pKa = 11.84PKK75 pKa = 10.3RR76 pKa = 11.84LRR78 pKa = 11.84LIGDD82 pKa = 4.53PYY84 pKa = 11.32LDD86 pKa = 4.08PCDD89 pKa = 4.92GIVGKK94 pKa = 10.34ILDD97 pKa = 4.49DD98 pKa = 3.7CTGLVQEE105 pKa = 5.53VVIQPKK111 pKa = 6.53WWRR114 pKa = 11.84FISMSSPTINQDD126 pKa = 2.86EE127 pKa = 4.45DD128 pKa = 3.89NEE130 pKa = 4.35CAILGNSYY138 pKa = 10.45SQVVPRR144 pKa = 11.84SEE146 pKa = 4.12PGSFVLLKK154 pKa = 10.93VNEE157 pKa = 4.28EE158 pKa = 4.29VVGAGCRR165 pKa = 11.84VIYY168 pKa = 10.68DD169 pKa = 3.58GGDD172 pKa = 3.51YY173 pKa = 10.98LLTAHH178 pKa = 6.67HH179 pKa = 6.75VWSQAPNHH187 pKa = 6.11IAKK190 pKa = 10.21GGKK193 pKa = 6.38TVEE196 pKa = 4.37ISMEE200 pKa = 4.06MKK202 pKa = 10.35PYY204 pKa = 10.5LSSKK208 pKa = 10.8NKK210 pKa = 9.77VLDD213 pKa = 3.73FCLVPVPAAVWSNLGVKK230 pKa = 9.69SSKK233 pKa = 9.96IASLHH238 pKa = 4.74QRR240 pKa = 11.84SNVTVYY246 pKa = 10.92GGTASTMLLSSFGIAEE262 pKa = 4.33ADD264 pKa = 3.79DD265 pKa = 3.82NPLRR269 pKa = 11.84IIHH272 pKa = 6.32KK273 pKa = 10.28ASTARR278 pKa = 11.84AWSGSPLYY286 pKa = 10.53NSNGLVLGVHH296 pKa = 6.95LGYY299 pKa = 10.72DD300 pKa = 3.49QLGSTNRR307 pKa = 11.84AVNIGYY313 pKa = 8.56VLRR316 pKa = 11.84TTSSNEE322 pKa = 3.68TAPPDD327 pKa = 3.72LNFVEE332 pKa = 4.52ITEE335 pKa = 4.61DD336 pKa = 3.55EE337 pKa = 4.39AVDD340 pKa = 3.73RR341 pKa = 11.84PSFDD345 pKa = 3.15EE346 pKa = 4.25YY347 pKa = 10.87EE348 pKa = 3.88IEE350 pKa = 4.23GFGKK354 pKa = 9.62IRR356 pKa = 11.84TRR358 pKa = 11.84GRR360 pKa = 11.84EE361 pKa = 3.65YY362 pKa = 10.75YY363 pKa = 9.97IPRR366 pKa = 11.84NKK368 pKa = 10.18DD369 pKa = 2.49WNKK372 pKa = 10.5YY373 pKa = 9.92DD374 pKa = 6.0DD375 pKa = 4.67EE376 pKa = 7.48DD377 pKa = 5.96DD378 pKa = 4.58DD379 pKa = 5.72AFFDD383 pKa = 4.22VPVALWLNSNEE394 pKa = 4.25TVEE397 pKa = 5.68KK398 pKa = 10.31PFKK401 pKa = 9.83LQRR404 pKa = 11.84GRR406 pKa = 11.84KK407 pKa = 6.72FTALAALIEE416 pKa = 4.33LGNYY420 pKa = 7.4SWEE423 pKa = 4.5SGGHH427 pKa = 4.66YY428 pKa = 9.31PQGVGLQFVGRR439 pKa = 11.84STCLFRR445 pKa = 11.84EE446 pKa = 4.49SSRR449 pKa = 11.84KK450 pKa = 9.55ALSEE454 pKa = 3.93RR455 pKa = 11.84FKK457 pKa = 10.93LASEE461 pKa = 4.71TFPEE465 pKa = 4.19LLDD468 pKa = 4.04YY469 pKa = 10.74NWPARR474 pKa = 11.84GSKK477 pKa = 10.8AEE479 pKa = 4.36LGSLLLQAEE488 pKa = 4.87RR489 pKa = 11.84FRR491 pKa = 11.84CTEE494 pKa = 3.73PPEE497 pKa = 4.34NLASCCDD504 pKa = 3.85KK505 pKa = 10.88LASRR509 pKa = 11.84YY510 pKa = 8.47PVTRR514 pKa = 11.84PRR516 pKa = 11.84CCFRR520 pKa = 11.84TDD522 pKa = 2.62KK523 pKa = 10.42WSWSAVEE530 pKa = 4.41EE531 pKa = 4.23EE532 pKa = 4.77VKK534 pKa = 10.69RR535 pKa = 11.84LAAQGRR541 pKa = 11.84EE542 pKa = 4.14VCKK545 pKa = 10.57DD546 pKa = 3.52SSPGSPLASLCKK558 pKa = 10.14RR559 pKa = 11.84NQDD562 pKa = 3.15VLAAHH567 pKa = 7.36LDD569 pKa = 3.96FVVKK573 pKa = 10.49AVTEE577 pKa = 4.12RR578 pKa = 11.84LFLLAEE584 pKa = 4.35TEE586 pKa = 4.18LHH588 pKa = 6.02GLSPVEE594 pKa = 5.59LINQGCCDD602 pKa = 3.88PVRR605 pKa = 11.84LFVKK609 pKa = 10.17QEE611 pKa = 3.45PHH613 pKa = 4.77TLKK616 pKa = 10.65KK617 pKa = 10.33INEE620 pKa = 3.7GRR622 pKa = 11.84YY623 pKa = 9.14RR624 pKa = 11.84LISSVSLVDD633 pKa = 3.76QIVEE637 pKa = 4.03RR638 pKa = 11.84MLFGPQNRR646 pKa = 11.84AEE648 pKa = 3.95IALWDD653 pKa = 4.54TIPSKK658 pKa = 10.8PGMGLTLKK666 pKa = 10.53SQARR670 pKa = 11.84KK671 pKa = 9.82IFGDD675 pKa = 3.67LLVKK679 pKa = 9.39HH680 pKa = 5.69THH682 pKa = 5.85CPAYY686 pKa = 9.85EE687 pKa = 4.0ADD689 pKa = 3.17ISGFDD694 pKa = 3.33WTVQDD699 pKa = 3.38WEE701 pKa = 4.17LWADD705 pKa = 3.26VEE707 pKa = 5.27IRR709 pKa = 11.84IKK711 pKa = 10.79LCSAGVNLAQCMRR724 pKa = 11.84NRR726 pKa = 11.84FYY728 pKa = 11.53CFMNSVFQLSDD739 pKa = 2.97GTLIQQCSPGVMKK752 pKa = 10.69SGSYY756 pKa = 8.6CTSSTNSRR764 pKa = 11.84VRR766 pKa = 11.84CLMAEE771 pKa = 4.45IIGSPWCIAMGDD783 pKa = 3.95DD784 pKa = 3.6SVEE787 pKa = 4.34GYY789 pKa = 10.66VEE791 pKa = 4.33GARR794 pKa = 11.84DD795 pKa = 3.35KK796 pKa = 11.56YY797 pKa = 11.08DD798 pKa = 3.89ALGHH802 pKa = 5.92KK803 pKa = 10.31CKK805 pKa = 10.39DD806 pKa = 3.66YY807 pKa = 10.96QVCDD811 pKa = 3.26SDD813 pKa = 4.74GLLLRR818 pKa = 11.84SVGFCSHH825 pKa = 7.18HH826 pKa = 6.66IDD828 pKa = 3.87SSGAYY833 pKa = 6.7LTSWAKK839 pKa = 9.3TLFKK843 pKa = 10.47HH844 pKa = 6.31LHH846 pKa = 6.02SKK848 pKa = 11.15DD849 pKa = 3.6EE850 pKa = 4.36DD851 pKa = 3.8FQDD854 pKa = 4.14IEE856 pKa = 4.2IEE858 pKa = 4.18LRR860 pKa = 11.84NSPMWPSIRR869 pKa = 11.84RR870 pKa = 11.84YY871 pKa = 9.38LVQEE875 pKa = 4.19TPSLDD880 pKa = 3.19KK881 pKa = 10.67RR882 pKa = 11.84YY883 pKa = 10.01EE884 pKa = 4.0PNEE887 pKa = 3.92EE888 pKa = 4.27EE889 pKa = 4.16EE890 pKa = 4.48VQTTEE895 pKa = 4.0QAVKK899 pKa = 9.28STAQQVHH906 pKa = 6.05GASCFTTCCEE916 pKa = 4.13RR917 pKa = 11.84NCYY920 pKa = 8.45GQGYY924 pKa = 10.29ACVQWW929 pKa = 3.84
MM1 pKa = 7.15HH2 pKa = 7.85RR3 pKa = 11.84SPFGYY8 pKa = 10.5LLLLISLPAGAVTYY22 pKa = 10.62DD23 pKa = 4.78CIRR26 pKa = 11.84GDD28 pKa = 3.99CQAGPNSLMMALMTLLTTGMWWVVSLSLFWIEE60 pKa = 3.78RR61 pKa = 11.84QRR63 pKa = 11.84YY64 pKa = 8.18RR65 pKa = 11.84SAEE68 pKa = 4.05EE69 pKa = 4.11KK70 pKa = 10.04ISRR73 pKa = 11.84PKK75 pKa = 10.3RR76 pKa = 11.84LRR78 pKa = 11.84LIGDD82 pKa = 4.53PYY84 pKa = 11.32LDD86 pKa = 4.08PCDD89 pKa = 4.92GIVGKK94 pKa = 10.34ILDD97 pKa = 4.49DD98 pKa = 3.7CTGLVQEE105 pKa = 5.53VVIQPKK111 pKa = 6.53WWRR114 pKa = 11.84FISMSSPTINQDD126 pKa = 2.86EE127 pKa = 4.45DD128 pKa = 3.89NEE130 pKa = 4.35CAILGNSYY138 pKa = 10.45SQVVPRR144 pKa = 11.84SEE146 pKa = 4.12PGSFVLLKK154 pKa = 10.93VNEE157 pKa = 4.28EE158 pKa = 4.29VVGAGCRR165 pKa = 11.84VIYY168 pKa = 10.68DD169 pKa = 3.58GGDD172 pKa = 3.51YY173 pKa = 10.98LLTAHH178 pKa = 6.67HH179 pKa = 6.75VWSQAPNHH187 pKa = 6.11IAKK190 pKa = 10.21GGKK193 pKa = 6.38TVEE196 pKa = 4.37ISMEE200 pKa = 4.06MKK202 pKa = 10.35PYY204 pKa = 10.5LSSKK208 pKa = 10.8NKK210 pKa = 9.77VLDD213 pKa = 3.73FCLVPVPAAVWSNLGVKK230 pKa = 9.69SSKK233 pKa = 9.96IASLHH238 pKa = 4.74QRR240 pKa = 11.84SNVTVYY246 pKa = 10.92GGTASTMLLSSFGIAEE262 pKa = 4.33ADD264 pKa = 3.79DD265 pKa = 3.82NPLRR269 pKa = 11.84IIHH272 pKa = 6.32KK273 pKa = 10.28ASTARR278 pKa = 11.84AWSGSPLYY286 pKa = 10.53NSNGLVLGVHH296 pKa = 6.95LGYY299 pKa = 10.72DD300 pKa = 3.49QLGSTNRR307 pKa = 11.84AVNIGYY313 pKa = 8.56VLRR316 pKa = 11.84TTSSNEE322 pKa = 3.68TAPPDD327 pKa = 3.72LNFVEE332 pKa = 4.52ITEE335 pKa = 4.61DD336 pKa = 3.55EE337 pKa = 4.39AVDD340 pKa = 3.73RR341 pKa = 11.84PSFDD345 pKa = 3.15EE346 pKa = 4.25YY347 pKa = 10.87EE348 pKa = 3.88IEE350 pKa = 4.23GFGKK354 pKa = 9.62IRR356 pKa = 11.84TRR358 pKa = 11.84GRR360 pKa = 11.84EE361 pKa = 3.65YY362 pKa = 10.75YY363 pKa = 9.97IPRR366 pKa = 11.84NKK368 pKa = 10.18DD369 pKa = 2.49WNKK372 pKa = 10.5YY373 pKa = 9.92DD374 pKa = 6.0DD375 pKa = 4.67EE376 pKa = 7.48DD377 pKa = 5.96DD378 pKa = 4.58DD379 pKa = 5.72AFFDD383 pKa = 4.22VPVALWLNSNEE394 pKa = 4.25TVEE397 pKa = 5.68KK398 pKa = 10.31PFKK401 pKa = 9.83LQRR404 pKa = 11.84GRR406 pKa = 11.84KK407 pKa = 6.72FTALAALIEE416 pKa = 4.33LGNYY420 pKa = 7.4SWEE423 pKa = 4.5SGGHH427 pKa = 4.66YY428 pKa = 9.31PQGVGLQFVGRR439 pKa = 11.84STCLFRR445 pKa = 11.84EE446 pKa = 4.49SSRR449 pKa = 11.84KK450 pKa = 9.55ALSEE454 pKa = 3.93RR455 pKa = 11.84FKK457 pKa = 10.93LASEE461 pKa = 4.71TFPEE465 pKa = 4.19LLDD468 pKa = 4.04YY469 pKa = 10.74NWPARR474 pKa = 11.84GSKK477 pKa = 10.8AEE479 pKa = 4.36LGSLLLQAEE488 pKa = 4.87RR489 pKa = 11.84FRR491 pKa = 11.84CTEE494 pKa = 3.73PPEE497 pKa = 4.34NLASCCDD504 pKa = 3.85KK505 pKa = 10.88LASRR509 pKa = 11.84YY510 pKa = 8.47PVTRR514 pKa = 11.84PRR516 pKa = 11.84CCFRR520 pKa = 11.84TDD522 pKa = 2.62KK523 pKa = 10.42WSWSAVEE530 pKa = 4.41EE531 pKa = 4.23EE532 pKa = 4.77VKK534 pKa = 10.69RR535 pKa = 11.84LAAQGRR541 pKa = 11.84EE542 pKa = 4.14VCKK545 pKa = 10.57DD546 pKa = 3.52SSPGSPLASLCKK558 pKa = 10.14RR559 pKa = 11.84NQDD562 pKa = 3.15VLAAHH567 pKa = 7.36LDD569 pKa = 3.96FVVKK573 pKa = 10.49AVTEE577 pKa = 4.12RR578 pKa = 11.84LFLLAEE584 pKa = 4.35TEE586 pKa = 4.18LHH588 pKa = 6.02GLSPVEE594 pKa = 5.59LINQGCCDD602 pKa = 3.88PVRR605 pKa = 11.84LFVKK609 pKa = 10.17QEE611 pKa = 3.45PHH613 pKa = 4.77TLKK616 pKa = 10.65KK617 pKa = 10.33INEE620 pKa = 3.7GRR622 pKa = 11.84YY623 pKa = 9.14RR624 pKa = 11.84LISSVSLVDD633 pKa = 3.76QIVEE637 pKa = 4.03RR638 pKa = 11.84MLFGPQNRR646 pKa = 11.84AEE648 pKa = 3.95IALWDD653 pKa = 4.54TIPSKK658 pKa = 10.8PGMGLTLKK666 pKa = 10.53SQARR670 pKa = 11.84KK671 pKa = 9.82IFGDD675 pKa = 3.67LLVKK679 pKa = 9.39HH680 pKa = 5.69THH682 pKa = 5.85CPAYY686 pKa = 9.85EE687 pKa = 4.0ADD689 pKa = 3.17ISGFDD694 pKa = 3.33WTVQDD699 pKa = 3.38WEE701 pKa = 4.17LWADD705 pKa = 3.26VEE707 pKa = 5.27IRR709 pKa = 11.84IKK711 pKa = 10.79LCSAGVNLAQCMRR724 pKa = 11.84NRR726 pKa = 11.84FYY728 pKa = 11.53CFMNSVFQLSDD739 pKa = 2.97GTLIQQCSPGVMKK752 pKa = 10.69SGSYY756 pKa = 8.6CTSSTNSRR764 pKa = 11.84VRR766 pKa = 11.84CLMAEE771 pKa = 4.45IIGSPWCIAMGDD783 pKa = 3.95DD784 pKa = 3.6SVEE787 pKa = 4.34GYY789 pKa = 10.66VEE791 pKa = 4.33GARR794 pKa = 11.84DD795 pKa = 3.35KK796 pKa = 11.56YY797 pKa = 11.08DD798 pKa = 3.89ALGHH802 pKa = 5.92KK803 pKa = 10.31CKK805 pKa = 10.39DD806 pKa = 3.66YY807 pKa = 10.96QVCDD811 pKa = 3.26SDD813 pKa = 4.74GLLLRR818 pKa = 11.84SVGFCSHH825 pKa = 7.18HH826 pKa = 6.66IDD828 pKa = 3.87SSGAYY833 pKa = 6.7LTSWAKK839 pKa = 9.3TLFKK843 pKa = 10.47HH844 pKa = 6.31LHH846 pKa = 6.02SKK848 pKa = 11.15DD849 pKa = 3.6EE850 pKa = 4.36DD851 pKa = 3.8FQDD854 pKa = 4.14IEE856 pKa = 4.2IEE858 pKa = 4.18LRR860 pKa = 11.84NSPMWPSIRR869 pKa = 11.84RR870 pKa = 11.84YY871 pKa = 9.38LVQEE875 pKa = 4.19TPSLDD880 pKa = 3.19KK881 pKa = 10.67RR882 pKa = 11.84YY883 pKa = 10.01EE884 pKa = 4.0PNEE887 pKa = 3.92EE888 pKa = 4.27EE889 pKa = 4.16EE890 pKa = 4.48VQTTEE895 pKa = 4.0QAVKK899 pKa = 9.28STAQQVHH906 pKa = 6.05GASCFTTCCEE916 pKa = 4.13RR917 pKa = 11.84NCYY920 pKa = 8.45GQGYY924 pKa = 10.29ACVQWW929 pKa = 3.84
Molecular weight: 104.21 kDa
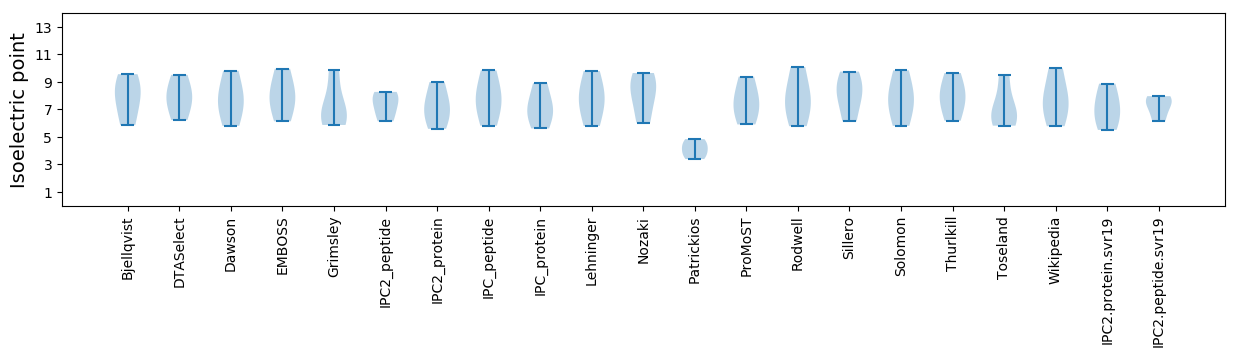
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|B5DC59|B5DC59_9VIRU Capsid protein OS=Sowbane mosaic virus OX=378833 GN=CP PE=3 SV=1
MM1 pKa = 7.13NQTKK5 pKa = 10.05KK6 pKa = 10.59KK7 pKa = 10.02KK8 pKa = 9.73SRR10 pKa = 11.84PRR12 pKa = 11.84NKK14 pKa = 9.61QSKK17 pKa = 8.31AQPNKK22 pKa = 9.06STAQVVSQPAAKK34 pKa = 9.89GIVMGRR40 pKa = 11.84AMPVFNGKK48 pKa = 10.15AGVLRR53 pKa = 11.84LRR55 pKa = 11.84HH56 pKa = 5.99HH57 pKa = 7.35EE58 pKa = 3.83ILQTIVSTTDD68 pKa = 2.54KK69 pKa = 9.96YY70 pKa = 8.9TVRR73 pKa = 11.84SVNLVPANFSWLSGVAVNFSKK94 pKa = 10.52WKK96 pKa = 7.86WHH98 pKa = 5.87ALNLVYY104 pKa = 10.73VPACSTSVQGNIDD117 pKa = 3.3MALQYY122 pKa = 10.87DD123 pKa = 4.4GNDD126 pKa = 3.29IPATTPEE133 pKa = 4.62EE134 pKa = 3.91LSVMYY139 pKa = 9.52GHH141 pKa = 6.74SGGPVWSGTAGCRR154 pKa = 11.84LLDD157 pKa = 3.81NATMSGRR164 pKa = 11.84GKK166 pKa = 9.7PPEE169 pKa = 4.9AIATIVDD176 pKa = 3.25VSKK179 pKa = 10.77FSKK182 pKa = 10.65SNYY185 pKa = 8.97LYY187 pKa = 10.74RR188 pKa = 11.84SGALSGDD195 pKa = 3.68SQTIYY200 pKa = 10.62CPCDD204 pKa = 3.21LQYY207 pKa = 11.51GVTAGITPTVGAIGYY222 pKa = 8.53IWATYY227 pKa = 8.91DD228 pKa = 3.24VEE230 pKa = 4.37MFEE233 pKa = 4.69PLPARR238 pKa = 11.84LNKK241 pKa = 10.21
MM1 pKa = 7.13NQTKK5 pKa = 10.05KK6 pKa = 10.59KK7 pKa = 10.02KK8 pKa = 9.73SRR10 pKa = 11.84PRR12 pKa = 11.84NKK14 pKa = 9.61QSKK17 pKa = 8.31AQPNKK22 pKa = 9.06STAQVVSQPAAKK34 pKa = 9.89GIVMGRR40 pKa = 11.84AMPVFNGKK48 pKa = 10.15AGVLRR53 pKa = 11.84LRR55 pKa = 11.84HH56 pKa = 5.99HH57 pKa = 7.35EE58 pKa = 3.83ILQTIVSTTDD68 pKa = 2.54KK69 pKa = 9.96YY70 pKa = 8.9TVRR73 pKa = 11.84SVNLVPANFSWLSGVAVNFSKK94 pKa = 10.52WKK96 pKa = 7.86WHH98 pKa = 5.87ALNLVYY104 pKa = 10.73VPACSTSVQGNIDD117 pKa = 3.3MALQYY122 pKa = 10.87DD123 pKa = 4.4GNDD126 pKa = 3.29IPATTPEE133 pKa = 4.62EE134 pKa = 3.91LSVMYY139 pKa = 9.52GHH141 pKa = 6.74SGGPVWSGTAGCRR154 pKa = 11.84LLDD157 pKa = 3.81NATMSGRR164 pKa = 11.84GKK166 pKa = 9.7PPEE169 pKa = 4.9AIATIVDD176 pKa = 3.25VSKK179 pKa = 10.77FSKK182 pKa = 10.65SNYY185 pKa = 8.97LYY187 pKa = 10.74RR188 pKa = 11.84SGALSGDD195 pKa = 3.68SQTIYY200 pKa = 10.62CPCDD204 pKa = 3.21LQYY207 pKa = 11.51GVTAGITPTVGAIGYY222 pKa = 8.53IWATYY227 pKa = 8.91DD228 pKa = 3.24VEE230 pKa = 4.37MFEE233 pKa = 4.69PLPARR238 pKa = 11.84LNKK241 pKa = 10.21
Molecular weight: 26.16 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1886 |
164 |
929 |
471.5 |
52.27 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.787 ± 0.401 | 3.022 ± 0.809 |
5.143 ± 0.623 | 5.62 ± 0.818 |
3.128 ± 0.439 | 7.052 ± 0.471 |
1.856 ± 0.158 | 4.613 ± 0.103 |
5.355 ± 0.366 | 8.961 ± 1.114 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.803 ± 0.252 | 4.083 ± 0.319 |
5.514 ± 0.476 | 3.712 ± 0.176 |
5.514 ± 0.372 | 9.544 ± 0.516 |
5.143 ± 0.552 | 7.635 ± 0.797 |
2.121 ± 0.15 | 3.393 ± 0.174 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
