
Fly associated circular virus 4
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cremevirales; Smacoviridae; Porprismacovirus; Porprismacovirus flas1
Average proteome isoelectric point is 6.76
Get precalculated fractions of proteins
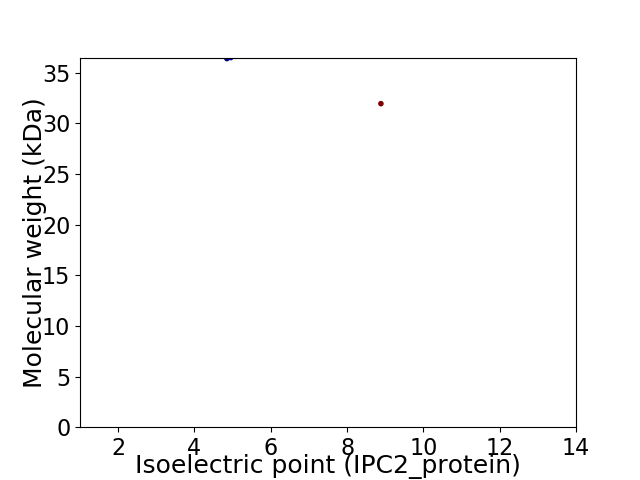
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A346BPB5|A0A346BPB5_9VIRU Putative capsid protein OS=Fly associated circular virus 4 OX=2293284 PE=4 SV=1
MM1 pKa = 7.21FVQVSEE7 pKa = 4.54TYY9 pKa = 10.7DD10 pKa = 3.44LSTQIGKK17 pKa = 8.67MGFVAIHH24 pKa = 6.09TPDD27 pKa = 3.21LQLIKK32 pKa = 10.9KK33 pKa = 6.71MWKK36 pKa = 9.37GLLMNHH42 pKa = 7.22RR43 pKa = 11.84YY44 pKa = 9.96YY45 pKa = 11.15KK46 pKa = 10.54FQSCDD51 pKa = 2.92LAMACASMLPADD63 pKa = 4.27PLQVGVEE70 pKa = 4.12SGEE73 pKa = 4.09IAPQDD78 pKa = 3.59MFNSILYY85 pKa = 9.77RR86 pKa = 11.84AVSNDD91 pKa = 3.03SYY93 pKa = 11.33NTILNRR99 pKa = 11.84IQNKK103 pKa = 7.58YY104 pKa = 8.34QSPVSGVDD112 pKa = 3.29KK113 pKa = 11.23NSVISDD119 pKa = 3.78NSDD122 pKa = 2.96SLADD126 pKa = 3.45VDD128 pKa = 5.81QFDD131 pKa = 4.89LYY133 pKa = 11.28YY134 pKa = 11.12GLLAEE139 pKa = 4.27PSGWRR144 pKa = 11.84KK145 pKa = 10.0AMPQNGLQMSNLYY158 pKa = 10.37PMVFQLVSNVGNNTGANTGSAFSLSKK184 pKa = 10.37LNSMFSSPGLGDD196 pKa = 3.95DD197 pKa = 3.44QASAGGVVQYY207 pKa = 11.87AMFRR211 pKa = 11.84GPSMRR216 pKa = 11.84MPRR219 pKa = 11.84IPTFGISSTSSEE231 pKa = 4.22IIGDD235 pKa = 3.77DD236 pKa = 3.23VTAIGEE242 pKa = 4.31FQSTPITYY250 pKa = 8.6VACIVLPPAKK260 pKa = 10.49LNVLYY265 pKa = 10.86YY266 pKa = 9.86RR267 pKa = 11.84LKK269 pKa = 9.03VTWTIEE275 pKa = 3.79FSEE278 pKa = 5.2PYY280 pKa = 9.48PATEE284 pKa = 3.57IANYY288 pKa = 9.64TSIANIGNQSYY299 pKa = 10.88GSDD302 pKa = 3.41YY303 pKa = 11.05DD304 pKa = 3.66AQSNFMSVKK313 pKa = 9.53TSSVDD318 pKa = 3.26AKK320 pKa = 10.62DD321 pKa = 4.53ANVQKK326 pKa = 10.69IMEE329 pKa = 4.48SGRR332 pKa = 3.55
MM1 pKa = 7.21FVQVSEE7 pKa = 4.54TYY9 pKa = 10.7DD10 pKa = 3.44LSTQIGKK17 pKa = 8.67MGFVAIHH24 pKa = 6.09TPDD27 pKa = 3.21LQLIKK32 pKa = 10.9KK33 pKa = 6.71MWKK36 pKa = 9.37GLLMNHH42 pKa = 7.22RR43 pKa = 11.84YY44 pKa = 9.96YY45 pKa = 11.15KK46 pKa = 10.54FQSCDD51 pKa = 2.92LAMACASMLPADD63 pKa = 4.27PLQVGVEE70 pKa = 4.12SGEE73 pKa = 4.09IAPQDD78 pKa = 3.59MFNSILYY85 pKa = 9.77RR86 pKa = 11.84AVSNDD91 pKa = 3.03SYY93 pKa = 11.33NTILNRR99 pKa = 11.84IQNKK103 pKa = 7.58YY104 pKa = 8.34QSPVSGVDD112 pKa = 3.29KK113 pKa = 11.23NSVISDD119 pKa = 3.78NSDD122 pKa = 2.96SLADD126 pKa = 3.45VDD128 pKa = 5.81QFDD131 pKa = 4.89LYY133 pKa = 11.28YY134 pKa = 11.12GLLAEE139 pKa = 4.27PSGWRR144 pKa = 11.84KK145 pKa = 10.0AMPQNGLQMSNLYY158 pKa = 10.37PMVFQLVSNVGNNTGANTGSAFSLSKK184 pKa = 10.37LNSMFSSPGLGDD196 pKa = 3.95DD197 pKa = 3.44QASAGGVVQYY207 pKa = 11.87AMFRR211 pKa = 11.84GPSMRR216 pKa = 11.84MPRR219 pKa = 11.84IPTFGISSTSSEE231 pKa = 4.22IIGDD235 pKa = 3.77DD236 pKa = 3.23VTAIGEE242 pKa = 4.31FQSTPITYY250 pKa = 8.6VACIVLPPAKK260 pKa = 10.49LNVLYY265 pKa = 10.86YY266 pKa = 9.86RR267 pKa = 11.84LKK269 pKa = 9.03VTWTIEE275 pKa = 3.79FSEE278 pKa = 5.2PYY280 pKa = 9.48PATEE284 pKa = 3.57IANYY288 pKa = 9.64TSIANIGNQSYY299 pKa = 10.88GSDD302 pKa = 3.41YY303 pKa = 11.05DD304 pKa = 3.66AQSNFMSVKK313 pKa = 9.53TSSVDD318 pKa = 3.26AKK320 pKa = 10.62DD321 pKa = 4.53ANVQKK326 pKa = 10.69IMEE329 pKa = 4.48SGRR332 pKa = 3.55
Molecular weight: 36.38 kDa
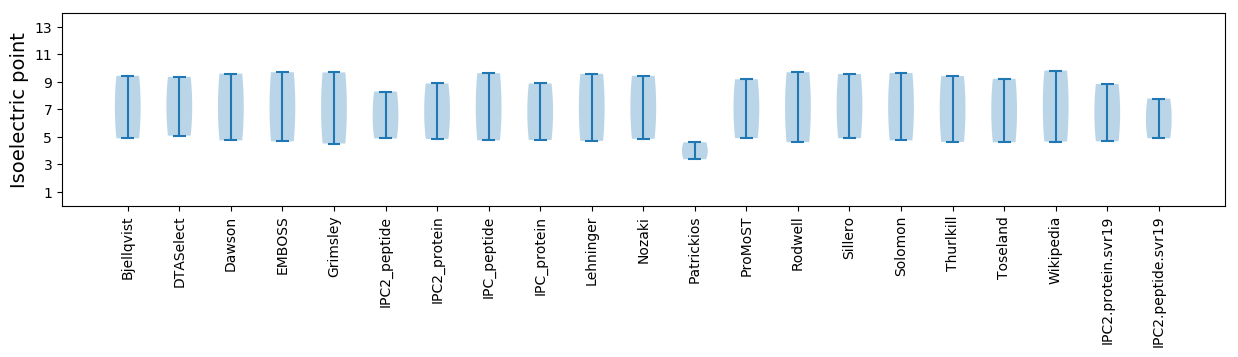
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A346BPB5|A0A346BPB5_9VIRU Putative capsid protein OS=Fly associated circular virus 4 OX=2293284 PE=4 SV=1
MM1 pKa = 7.06VKK3 pKa = 10.33VYY5 pKa = 10.77LLTVPLVHH13 pKa = 7.45RR14 pKa = 11.84IQFDD18 pKa = 3.57SPLYY22 pKa = 8.97RR23 pKa = 11.84WRR25 pKa = 11.84HH26 pKa = 4.54SSSYY30 pKa = 7.17STKK33 pKa = 9.21RR34 pKa = 11.84AIRR37 pKa = 11.84ILIEE41 pKa = 4.16SLDD44 pKa = 3.78CKK46 pKa = 10.21RR47 pKa = 11.84WIVARR52 pKa = 11.84EE53 pKa = 3.87RR54 pKa = 11.84GRR56 pKa = 11.84GGYY59 pKa = 8.2EE60 pKa = 3.53HH61 pKa = 6.8LQVRR65 pKa = 11.84VEE67 pKa = 4.19TSADD71 pKa = 3.26NFFDD75 pKa = 3.43RR76 pKa = 11.84VSRR79 pKa = 11.84VLPCAHH85 pKa = 6.74IEE87 pKa = 4.17EE88 pKa = 4.78STSAYY93 pKa = 10.08SDD95 pKa = 2.73WYY97 pKa = 8.95EE98 pKa = 4.14RR99 pKa = 11.84KK100 pKa = 8.93EE101 pKa = 3.93GRR103 pKa = 11.84YY104 pKa = 7.19VTSSDD109 pKa = 3.83LLPGRR114 pKa = 11.84TKK116 pKa = 10.69VLKK119 pKa = 10.32QRR121 pKa = 11.84FGCFYY126 pKa = 9.86PFQKK130 pKa = 10.23RR131 pKa = 11.84VLRR134 pKa = 11.84ALQSTNDD141 pKa = 3.49RR142 pKa = 11.84EE143 pKa = 4.44VVVWYY148 pKa = 9.56DD149 pKa = 3.2QTGNAGKK156 pKa = 9.92SWLKK160 pKa = 9.6GALWEE165 pKa = 4.73RR166 pKa = 11.84GLAYY170 pKa = 9.62IIQPMSSISVMVQDD184 pKa = 4.78VADD187 pKa = 4.02EE188 pKa = 4.25YY189 pKa = 11.32LRR191 pKa = 11.84HH192 pKa = 6.82GYY194 pKa = 9.01RR195 pKa = 11.84QYY197 pKa = 11.69LVIDD201 pKa = 4.22IPRR204 pKa = 11.84TWKK207 pKa = 9.79WSEE210 pKa = 4.01DD211 pKa = 3.82LYY213 pKa = 11.44CAIEE217 pKa = 4.27TIKK220 pKa = 11.03DD221 pKa = 3.63GLLKK225 pKa = 10.54DD226 pKa = 3.47PRR228 pKa = 11.84YY229 pKa = 10.44NSEE232 pKa = 4.03TVNIAGVQVLILCNTLPKK250 pKa = 9.99LDD252 pKa = 4.24KK253 pKa = 10.94LSVDD257 pKa = 2.76RR258 pKa = 11.84WIILNRR264 pKa = 11.84DD265 pKa = 3.19GTRR268 pKa = 11.84RR269 pKa = 11.84GSFSS273 pKa = 3.05
MM1 pKa = 7.06VKK3 pKa = 10.33VYY5 pKa = 10.77LLTVPLVHH13 pKa = 7.45RR14 pKa = 11.84IQFDD18 pKa = 3.57SPLYY22 pKa = 8.97RR23 pKa = 11.84WRR25 pKa = 11.84HH26 pKa = 4.54SSSYY30 pKa = 7.17STKK33 pKa = 9.21RR34 pKa = 11.84AIRR37 pKa = 11.84ILIEE41 pKa = 4.16SLDD44 pKa = 3.78CKK46 pKa = 10.21RR47 pKa = 11.84WIVARR52 pKa = 11.84EE53 pKa = 3.87RR54 pKa = 11.84GRR56 pKa = 11.84GGYY59 pKa = 8.2EE60 pKa = 3.53HH61 pKa = 6.8LQVRR65 pKa = 11.84VEE67 pKa = 4.19TSADD71 pKa = 3.26NFFDD75 pKa = 3.43RR76 pKa = 11.84VSRR79 pKa = 11.84VLPCAHH85 pKa = 6.74IEE87 pKa = 4.17EE88 pKa = 4.78STSAYY93 pKa = 10.08SDD95 pKa = 2.73WYY97 pKa = 8.95EE98 pKa = 4.14RR99 pKa = 11.84KK100 pKa = 8.93EE101 pKa = 3.93GRR103 pKa = 11.84YY104 pKa = 7.19VTSSDD109 pKa = 3.83LLPGRR114 pKa = 11.84TKK116 pKa = 10.69VLKK119 pKa = 10.32QRR121 pKa = 11.84FGCFYY126 pKa = 9.86PFQKK130 pKa = 10.23RR131 pKa = 11.84VLRR134 pKa = 11.84ALQSTNDD141 pKa = 3.49RR142 pKa = 11.84EE143 pKa = 4.44VVVWYY148 pKa = 9.56DD149 pKa = 3.2QTGNAGKK156 pKa = 9.92SWLKK160 pKa = 9.6GALWEE165 pKa = 4.73RR166 pKa = 11.84GLAYY170 pKa = 9.62IIQPMSSISVMVQDD184 pKa = 4.78VADD187 pKa = 4.02EE188 pKa = 4.25YY189 pKa = 11.32LRR191 pKa = 11.84HH192 pKa = 6.82GYY194 pKa = 9.01RR195 pKa = 11.84QYY197 pKa = 11.69LVIDD201 pKa = 4.22IPRR204 pKa = 11.84TWKK207 pKa = 9.79WSEE210 pKa = 4.01DD211 pKa = 3.82LYY213 pKa = 11.44CAIEE217 pKa = 4.27TIKK220 pKa = 11.03DD221 pKa = 3.63GLLKK225 pKa = 10.54DD226 pKa = 3.47PRR228 pKa = 11.84YY229 pKa = 10.44NSEE232 pKa = 4.03TVNIAGVQVLILCNTLPKK250 pKa = 9.99LDD252 pKa = 4.24KK253 pKa = 10.94LSVDD257 pKa = 2.76RR258 pKa = 11.84WIILNRR264 pKa = 11.84DD265 pKa = 3.19GTRR268 pKa = 11.84RR269 pKa = 11.84GSFSS273 pKa = 3.05
Molecular weight: 31.94 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
605 |
273 |
332 |
302.5 |
34.16 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.95 ± 1.015 | 1.322 ± 0.332 |
6.116 ± 0.073 | 3.967 ± 0.758 |
3.306 ± 0.484 | 6.281 ± 0.513 |
1.157 ± 0.44 | 6.116 ± 0.073 |
4.628 ± 0.326 | 8.43 ± 0.953 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.14 ± 1.332 | 4.628 ± 1.347 |
4.298 ± 0.653 | 4.628 ± 0.63 |
6.116 ± 2.702 | 10.248 ± 1.19 |
4.793 ± 0.021 | 7.603 ± 0.536 |
1.983 ± 0.857 | 5.289 ± 0.134 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
