
Eggerthella sp. CAG:298
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Coriobacteriia; Eggerthellales; Eggerthellaceae; Eggerthella; environmental samples
Average proteome isoelectric point is 5.65
Get precalculated fractions of proteins
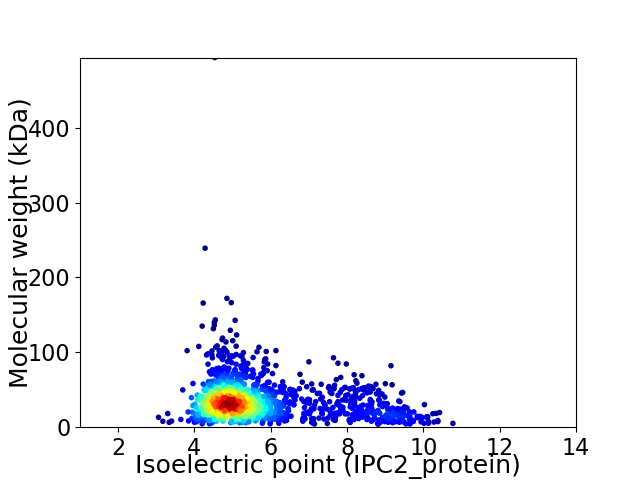
Virtual 2D-PAGE plot for 1581 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R7B5E6|R7B5E6_9ACTN CRISPR-associated endonuclease Cas1 OS=Eggerthella sp. CAG:298 OX=1262876 GN=cas1 PE=3 SV=1
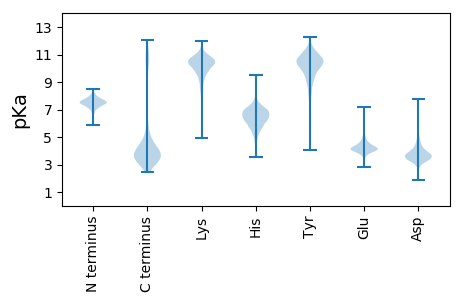
MM1 pKa = 7.43KK2 pKa = 10.21KK3 pKa = 9.77ISKK6 pKa = 9.95AKK8 pKa = 10.08NSFVAHH14 pKa = 6.94AIVVAVSLFLVAFSLSAPGVAFAEE38 pKa = 4.33DD39 pKa = 3.94VEE41 pKa = 4.79VQSSEE46 pKa = 4.0PTQTLIDD53 pKa = 3.9QPAVAAEE60 pKa = 3.85ATPAVEE66 pKa = 4.15AVSVPSDD73 pKa = 3.39PAQVSGVEE81 pKa = 4.02AVPGNSSTQKK91 pKa = 10.65SEE93 pKa = 4.14GASSALNNDD102 pKa = 2.78KK103 pKa = 9.28NTNVEE108 pKa = 4.08NSASDD113 pKa = 3.56PAASNDD119 pKa = 3.49SNATEE124 pKa = 4.58FSTTQVEE131 pKa = 4.65EE132 pKa = 3.99KK133 pKa = 10.43ATIKK137 pKa = 10.26PAADD141 pKa = 3.27SPHH144 pKa = 6.27RR145 pKa = 11.84VNLSFKK151 pKa = 10.25ILYY154 pKa = 9.28QGQEE158 pKa = 3.82FTDD161 pKa = 5.09GYY163 pKa = 10.81EE164 pKa = 4.01IGHH167 pKa = 6.28SDD169 pKa = 3.3SVSFVCTMGDD179 pKa = 3.28SHH181 pKa = 7.79ASTAASHH188 pKa = 6.48MISMGDD194 pKa = 3.39VLIEE198 pKa = 3.88RR199 pKa = 11.84DD200 pKa = 3.65ALAKK204 pKa = 9.83FLRR207 pKa = 11.84EE208 pKa = 4.16GYY210 pKa = 9.75EE211 pKa = 3.46IRR213 pKa = 11.84GWTKK217 pKa = 10.74SLLAGAEE224 pKa = 4.16VYY226 pKa = 10.87GFDD229 pKa = 4.81RR230 pKa = 11.84DD231 pKa = 4.72GIEE234 pKa = 4.36CMDD237 pKa = 4.16GEE239 pKa = 4.73TIYY242 pKa = 10.48IVATYY247 pKa = 10.42GPAANQFTVNFDD259 pKa = 3.32AMGGSNAPEE268 pKa = 4.23SLTEE272 pKa = 3.78VSTEE276 pKa = 3.65NEE278 pKa = 4.1YY279 pKa = 9.71TFTISDD285 pKa = 3.8AVPTRR290 pKa = 11.84EE291 pKa = 4.2GYY293 pKa = 10.94NFVGWSTNSSLLPPYY308 pKa = 10.18LKK310 pKa = 10.84AGDD313 pKa = 3.67TVTVSEE319 pKa = 4.27NDD321 pKa = 3.24SYY323 pKa = 11.89AITLYY328 pKa = 10.87AVWEE332 pKa = 4.12IANKK336 pKa = 10.05VEE338 pKa = 4.0LVYY341 pKa = 10.95AVDD344 pKa = 3.9YY345 pKa = 10.91VQFGDD350 pKa = 4.02AEE352 pKa = 4.6TYY354 pKa = 10.39AQGSSAVVKK363 pKa = 10.44GCDD366 pKa = 3.65YY367 pKa = 11.31VKK369 pKa = 10.56EE370 pKa = 4.49GYY372 pKa = 10.42KK373 pKa = 10.52FIGWSLDD380 pKa = 3.14PMSDD384 pKa = 2.85HH385 pKa = 7.37ASYY388 pKa = 11.29KK389 pKa = 10.45EE390 pKa = 3.88GDD392 pKa = 4.24SIIMDD397 pKa = 3.92STSGSIWLYY406 pKa = 9.52AVWVRR411 pKa = 11.84TYY413 pKa = 10.51TVTYY417 pKa = 10.02TDD419 pKa = 3.81GQGGTVFEE427 pKa = 4.32NQVISGVEE435 pKa = 4.19SGTSTPTFPVSRR447 pKa = 11.84PSSIVIDD454 pKa = 3.64GNTYY458 pKa = 10.21VFSGWDD464 pKa = 3.3NNAEE468 pKa = 4.17VVLGDD473 pKa = 4.1MIINAIWKK481 pKa = 9.78LDD483 pKa = 3.66ANGNGVADD491 pKa = 3.77EE492 pKa = 5.07DD493 pKa = 3.89EE494 pKa = 5.03AKK496 pKa = 10.84FSVTYY501 pKa = 9.59TDD503 pKa = 3.93GVGGTAFGDD512 pKa = 3.91YY513 pKa = 10.53VISNLLVNTATPRR526 pKa = 11.84YY527 pKa = 8.14RR528 pKa = 11.84APEE531 pKa = 3.55RR532 pKa = 11.84AGYY535 pKa = 9.47IFQGWSPTVADD546 pKa = 5.18IITGNAIYY554 pKa = 8.87TAIWAEE560 pKa = 4.02DD561 pKa = 3.52KK562 pKa = 11.27NNNNIPDD569 pKa = 4.21SEE571 pKa = 4.49EE572 pKa = 3.54EE573 pKa = 4.41TYY575 pKa = 10.58TIIFINDD582 pKa = 3.0VTGATLQTTPGVLGGLDD599 pKa = 3.48TPQFAGPAPTMTGYY613 pKa = 10.98VFVGWSPALAQTVDD627 pKa = 3.47PATADD632 pKa = 3.32EE633 pKa = 4.99FGNIYY638 pKa = 10.45YY639 pKa = 9.97IARR642 pKa = 11.84WEE644 pKa = 3.78VDD646 pKa = 2.99ANNNGIADD654 pKa = 3.75VDD656 pKa = 3.67EE657 pKa = 4.58ARR659 pKa = 11.84YY660 pKa = 9.94SVIYY664 pKa = 10.48SDD666 pKa = 4.02GANGSVFADD675 pKa = 3.39QTTTGLLSGLATPQFTGEE693 pKa = 4.1TPVRR697 pKa = 11.84SGYY700 pKa = 10.09IFAGWSSSPAGTIDD714 pKa = 3.3PTAADD719 pKa = 3.36SLGRR723 pKa = 11.84IIYY726 pKa = 6.66TALWAADD733 pKa = 3.51VNGNGVADD741 pKa = 4.1SEE743 pKa = 4.65EE744 pKa = 3.92PTYY747 pKa = 10.73TITYY751 pKa = 8.76TDD753 pKa = 3.94GANGAAFADD762 pKa = 4.13QVSSGLLGGVATPQFSGTPTRR783 pKa = 11.84TGYY786 pKa = 11.15KK787 pKa = 9.15FVGWSTEE794 pKa = 3.86PASTVSGDD802 pKa = 3.78VVYY805 pKa = 9.1TALWEE810 pKa = 4.2AVEE813 pKa = 4.42PDD815 pKa = 4.21PTPGTDD821 pKa = 5.1DD822 pKa = 3.58PTPTPGPDD830 pKa = 3.84DD831 pKa = 4.26PAPTPGDD838 pKa = 4.13DD839 pKa = 5.3DD840 pKa = 6.33DD841 pKa = 4.19NTGGNGSATTPGTNEE856 pKa = 4.33PGNVTPNPGPANGTTTPGEE875 pKa = 4.01TTIADD880 pKa = 4.29DD881 pKa = 4.06EE882 pKa = 4.79TPLAGGAGEE891 pKa = 4.6TEE893 pKa = 4.53DD894 pKa = 5.61DD895 pKa = 3.97PAEE898 pKa = 4.39GDD900 pKa = 3.9VADD903 pKa = 4.94DD904 pKa = 3.95AAVADD909 pKa = 4.6EE910 pKa = 4.32AAGAIDD916 pKa = 3.75EE917 pKa = 4.64AADD920 pKa = 3.72TEE922 pKa = 4.4ILDD925 pKa = 5.18DD926 pKa = 4.56EE927 pKa = 4.98NPLAAADD934 pKa = 4.78HH935 pKa = 6.07SSSWLSWLLTVGLLLVVCFGVFFLVRR961 pKa = 11.84RR962 pKa = 11.84RR963 pKa = 11.84KK964 pKa = 7.85NTRR967 pKa = 11.84EE968 pKa = 3.78QNQQ971 pKa = 3.04
MM1 pKa = 7.43KK2 pKa = 10.21KK3 pKa = 9.77ISKK6 pKa = 9.95AKK8 pKa = 10.08NSFVAHH14 pKa = 6.94AIVVAVSLFLVAFSLSAPGVAFAEE38 pKa = 4.33DD39 pKa = 3.94VEE41 pKa = 4.79VQSSEE46 pKa = 4.0PTQTLIDD53 pKa = 3.9QPAVAAEE60 pKa = 3.85ATPAVEE66 pKa = 4.15AVSVPSDD73 pKa = 3.39PAQVSGVEE81 pKa = 4.02AVPGNSSTQKK91 pKa = 10.65SEE93 pKa = 4.14GASSALNNDD102 pKa = 2.78KK103 pKa = 9.28NTNVEE108 pKa = 4.08NSASDD113 pKa = 3.56PAASNDD119 pKa = 3.49SNATEE124 pKa = 4.58FSTTQVEE131 pKa = 4.65EE132 pKa = 3.99KK133 pKa = 10.43ATIKK137 pKa = 10.26PAADD141 pKa = 3.27SPHH144 pKa = 6.27RR145 pKa = 11.84VNLSFKK151 pKa = 10.25ILYY154 pKa = 9.28QGQEE158 pKa = 3.82FTDD161 pKa = 5.09GYY163 pKa = 10.81EE164 pKa = 4.01IGHH167 pKa = 6.28SDD169 pKa = 3.3SVSFVCTMGDD179 pKa = 3.28SHH181 pKa = 7.79ASTAASHH188 pKa = 6.48MISMGDD194 pKa = 3.39VLIEE198 pKa = 3.88RR199 pKa = 11.84DD200 pKa = 3.65ALAKK204 pKa = 9.83FLRR207 pKa = 11.84EE208 pKa = 4.16GYY210 pKa = 9.75EE211 pKa = 3.46IRR213 pKa = 11.84GWTKK217 pKa = 10.74SLLAGAEE224 pKa = 4.16VYY226 pKa = 10.87GFDD229 pKa = 4.81RR230 pKa = 11.84DD231 pKa = 4.72GIEE234 pKa = 4.36CMDD237 pKa = 4.16GEE239 pKa = 4.73TIYY242 pKa = 10.48IVATYY247 pKa = 10.42GPAANQFTVNFDD259 pKa = 3.32AMGGSNAPEE268 pKa = 4.23SLTEE272 pKa = 3.78VSTEE276 pKa = 3.65NEE278 pKa = 4.1YY279 pKa = 9.71TFTISDD285 pKa = 3.8AVPTRR290 pKa = 11.84EE291 pKa = 4.2GYY293 pKa = 10.94NFVGWSTNSSLLPPYY308 pKa = 10.18LKK310 pKa = 10.84AGDD313 pKa = 3.67TVTVSEE319 pKa = 4.27NDD321 pKa = 3.24SYY323 pKa = 11.89AITLYY328 pKa = 10.87AVWEE332 pKa = 4.12IANKK336 pKa = 10.05VEE338 pKa = 4.0LVYY341 pKa = 10.95AVDD344 pKa = 3.9YY345 pKa = 10.91VQFGDD350 pKa = 4.02AEE352 pKa = 4.6TYY354 pKa = 10.39AQGSSAVVKK363 pKa = 10.44GCDD366 pKa = 3.65YY367 pKa = 11.31VKK369 pKa = 10.56EE370 pKa = 4.49GYY372 pKa = 10.42KK373 pKa = 10.52FIGWSLDD380 pKa = 3.14PMSDD384 pKa = 2.85HH385 pKa = 7.37ASYY388 pKa = 11.29KK389 pKa = 10.45EE390 pKa = 3.88GDD392 pKa = 4.24SIIMDD397 pKa = 3.92STSGSIWLYY406 pKa = 9.52AVWVRR411 pKa = 11.84TYY413 pKa = 10.51TVTYY417 pKa = 10.02TDD419 pKa = 3.81GQGGTVFEE427 pKa = 4.32NQVISGVEE435 pKa = 4.19SGTSTPTFPVSRR447 pKa = 11.84PSSIVIDD454 pKa = 3.64GNTYY458 pKa = 10.21VFSGWDD464 pKa = 3.3NNAEE468 pKa = 4.17VVLGDD473 pKa = 4.1MIINAIWKK481 pKa = 9.78LDD483 pKa = 3.66ANGNGVADD491 pKa = 3.77EE492 pKa = 5.07DD493 pKa = 3.89EE494 pKa = 5.03AKK496 pKa = 10.84FSVTYY501 pKa = 9.59TDD503 pKa = 3.93GVGGTAFGDD512 pKa = 3.91YY513 pKa = 10.53VISNLLVNTATPRR526 pKa = 11.84YY527 pKa = 8.14RR528 pKa = 11.84APEE531 pKa = 3.55RR532 pKa = 11.84AGYY535 pKa = 9.47IFQGWSPTVADD546 pKa = 5.18IITGNAIYY554 pKa = 8.87TAIWAEE560 pKa = 4.02DD561 pKa = 3.52KK562 pKa = 11.27NNNNIPDD569 pKa = 4.21SEE571 pKa = 4.49EE572 pKa = 3.54EE573 pKa = 4.41TYY575 pKa = 10.58TIIFINDD582 pKa = 3.0VTGATLQTTPGVLGGLDD599 pKa = 3.48TPQFAGPAPTMTGYY613 pKa = 10.98VFVGWSPALAQTVDD627 pKa = 3.47PATADD632 pKa = 3.32EE633 pKa = 4.99FGNIYY638 pKa = 10.45YY639 pKa = 9.97IARR642 pKa = 11.84WEE644 pKa = 3.78VDD646 pKa = 2.99ANNNGIADD654 pKa = 3.75VDD656 pKa = 3.67EE657 pKa = 4.58ARR659 pKa = 11.84YY660 pKa = 9.94SVIYY664 pKa = 10.48SDD666 pKa = 4.02GANGSVFADD675 pKa = 3.39QTTTGLLSGLATPQFTGEE693 pKa = 4.1TPVRR697 pKa = 11.84SGYY700 pKa = 10.09IFAGWSSSPAGTIDD714 pKa = 3.3PTAADD719 pKa = 3.36SLGRR723 pKa = 11.84IIYY726 pKa = 6.66TALWAADD733 pKa = 3.51VNGNGVADD741 pKa = 4.1SEE743 pKa = 4.65EE744 pKa = 3.92PTYY747 pKa = 10.73TITYY751 pKa = 8.76TDD753 pKa = 3.94GANGAAFADD762 pKa = 4.13QVSSGLLGGVATPQFSGTPTRR783 pKa = 11.84TGYY786 pKa = 11.15KK787 pKa = 9.15FVGWSTEE794 pKa = 3.86PASTVSGDD802 pKa = 3.78VVYY805 pKa = 9.1TALWEE810 pKa = 4.2AVEE813 pKa = 4.42PDD815 pKa = 4.21PTPGTDD821 pKa = 5.1DD822 pKa = 3.58PTPTPGPDD830 pKa = 3.84DD831 pKa = 4.26PAPTPGDD838 pKa = 4.13DD839 pKa = 5.3DD840 pKa = 6.33DD841 pKa = 4.19NTGGNGSATTPGTNEE856 pKa = 4.33PGNVTPNPGPANGTTTPGEE875 pKa = 4.01TTIADD880 pKa = 4.29DD881 pKa = 4.06EE882 pKa = 4.79TPLAGGAGEE891 pKa = 4.6TEE893 pKa = 4.53DD894 pKa = 5.61DD895 pKa = 3.97PAEE898 pKa = 4.39GDD900 pKa = 3.9VADD903 pKa = 4.94DD904 pKa = 3.95AAVADD909 pKa = 4.6EE910 pKa = 4.32AAGAIDD916 pKa = 3.75EE917 pKa = 4.64AADD920 pKa = 3.72TEE922 pKa = 4.4ILDD925 pKa = 5.18DD926 pKa = 4.56EE927 pKa = 4.98NPLAAADD934 pKa = 4.78HH935 pKa = 6.07SSSWLSWLLTVGLLLVVCFGVFFLVRR961 pKa = 11.84RR962 pKa = 11.84RR963 pKa = 11.84KK964 pKa = 7.85NTRR967 pKa = 11.84EE968 pKa = 3.78QNQQ971 pKa = 3.04
Molecular weight: 102.21 kDa
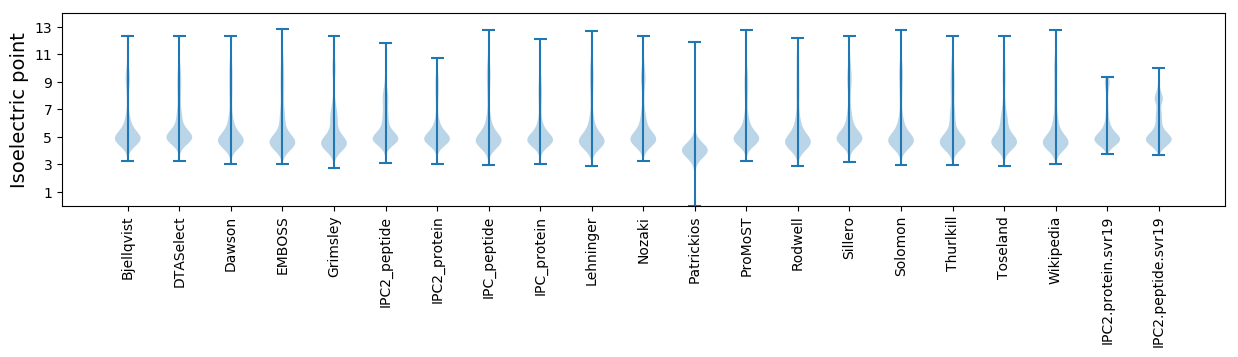
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R7B7V5|R7B7V5_9ACTN Membrane-flanked domain protein OS=Eggerthella sp. CAG:298 OX=1262876 GN=BN592_00099 PE=4 SV=1
MM1 pKa = 7.48LSKK4 pKa = 10.58DD5 pKa = 3.66ARR7 pKa = 11.84AQPMNRR13 pKa = 11.84SNPGVFDD20 pKa = 3.62HH21 pKa = 6.76EE22 pKa = 4.36RR23 pKa = 11.84LFIEE27 pKa = 4.88ALLLKK32 pKa = 10.24KK33 pKa = 9.22RR34 pKa = 11.84THH36 pKa = 6.08ARR38 pKa = 11.84FHH40 pKa = 6.43LSCTRR45 pKa = 11.84FAEE48 pKa = 4.85GSRR51 pKa = 11.84KK52 pKa = 9.81YY53 pKa = 10.69FIDD56 pKa = 5.26AIDD59 pKa = 3.47EE60 pKa = 4.67RR61 pKa = 11.84IFRR64 pKa = 11.84ARR66 pKa = 11.84FTHH69 pKa = 6.75
MM1 pKa = 7.48LSKK4 pKa = 10.58DD5 pKa = 3.66ARR7 pKa = 11.84AQPMNRR13 pKa = 11.84SNPGVFDD20 pKa = 3.62HH21 pKa = 6.76EE22 pKa = 4.36RR23 pKa = 11.84LFIEE27 pKa = 4.88ALLLKK32 pKa = 10.24KK33 pKa = 9.22RR34 pKa = 11.84THH36 pKa = 6.08ARR38 pKa = 11.84FHH40 pKa = 6.43LSCTRR45 pKa = 11.84FAEE48 pKa = 4.85GSRR51 pKa = 11.84KK52 pKa = 9.81YY53 pKa = 10.69FIDD56 pKa = 5.26AIDD59 pKa = 3.47EE60 pKa = 4.67RR61 pKa = 11.84IFRR64 pKa = 11.84ARR66 pKa = 11.84FTHH69 pKa = 6.75
Molecular weight: 8.22 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
518525 |
35 |
4679 |
328.0 |
35.98 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.414 ± 0.065 | 1.481 ± 0.028 |
5.883 ± 0.054 | 7.132 ± 0.069 |
4.029 ± 0.041 | 7.344 ± 0.06 |
1.978 ± 0.032 | 6.316 ± 0.053 |
4.513 ± 0.051 | 9.3 ± 0.082 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.577 ± 0.031 | 3.522 ± 0.043 |
4.179 ± 0.033 | 3.252 ± 0.029 |
5.076 ± 0.053 | 6.049 ± 0.054 |
5.552 ± 0.073 | 7.512 ± 0.054 |
0.939 ± 0.021 | 2.953 ± 0.031 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
