
Gynuella sunshinyii YC6258
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Oceanospirillales; Saccharospirillaceae; Gynuella; Gynuella sunshinyii
Average proteome isoelectric point is 6.23
Get precalculated fractions of proteins
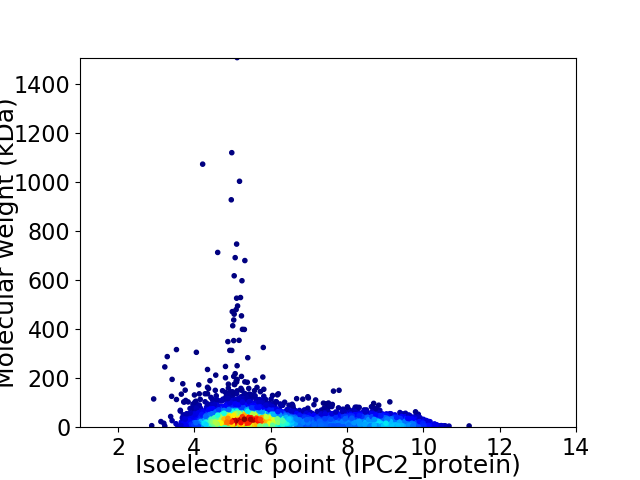
Virtual 2D-PAGE plot for 5913 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0C5VHJ8|A0A0C5VHJ8_9GAMM Uncharacterized protein OS=Gynuella sunshinyii YC6258 OX=1445510 GN=YC6258_02106 PE=4 SV=1
MM1 pKa = 7.14THH3 pKa = 6.4QKK5 pKa = 10.9LLLTAACFSIVTLTACNSSDD25 pKa = 4.98SGDD28 pKa = 3.34SDD30 pKa = 3.47NFKK33 pKa = 11.12RR34 pKa = 11.84NLQMNLSDD42 pKa = 3.87STTQALLDD50 pKa = 3.93DD51 pKa = 4.36TLATNAGTTTSLSIAQGDD69 pKa = 4.04LSADD73 pKa = 3.89EE74 pKa = 3.96ISIRR78 pKa = 11.84SLNNSSQVLARR89 pKa = 11.84KK90 pKa = 7.19MARR93 pKa = 11.84TAFNQNDD100 pKa = 3.37KK101 pKa = 10.58SACDD105 pKa = 3.45SGSVSVSISGDD116 pKa = 3.09IADD119 pKa = 5.46DD120 pKa = 3.72EE121 pKa = 4.78TSGNVTAALVASNCRR136 pKa = 11.84SYY138 pKa = 11.3EE139 pKa = 3.86SSDD142 pKa = 3.71GINIEE147 pKa = 3.98GMINGSLSDD156 pKa = 3.69SVSWQTSGSQFTALSSTSTGNLEE179 pKa = 3.51ITYY182 pKa = 10.46VGDD185 pKa = 3.92GSSDD189 pKa = 2.88GHH191 pKa = 6.1NYY193 pKa = 10.42SFGFLYY199 pKa = 10.76DD200 pKa = 4.82LSDD203 pKa = 4.31LNNSMSLNTVNNTLEE218 pKa = 3.62VDD220 pKa = 3.46ANYY223 pKa = 10.84SVYY226 pKa = 10.46IEE228 pKa = 4.57VYY230 pKa = 10.63YY231 pKa = 9.81EE232 pKa = 3.74TDD234 pKa = 3.1NDD236 pKa = 3.74KK237 pKa = 11.24EE238 pKa = 4.21RR239 pKa = 11.84VGGSMKK245 pKa = 10.4VATLDD250 pKa = 3.5SVIYY254 pKa = 9.59DD255 pKa = 3.7APTTTSSLSHH265 pKa = 6.5PVDD268 pKa = 3.06GRR270 pKa = 11.84IQVTAGGQTATVDD283 pKa = 4.67FDD285 pKa = 3.54QTDD288 pKa = 3.72YY289 pKa = 11.18TVTIGNVSNTYY300 pKa = 10.3DD301 pKa = 3.84YY302 pKa = 11.75EE303 pKa = 6.58DD304 pKa = 3.62DD305 pKa = 3.93TVFF308 pKa = 4.52
MM1 pKa = 7.14THH3 pKa = 6.4QKK5 pKa = 10.9LLLTAACFSIVTLTACNSSDD25 pKa = 4.98SGDD28 pKa = 3.34SDD30 pKa = 3.47NFKK33 pKa = 11.12RR34 pKa = 11.84NLQMNLSDD42 pKa = 3.87STTQALLDD50 pKa = 3.93DD51 pKa = 4.36TLATNAGTTTSLSIAQGDD69 pKa = 4.04LSADD73 pKa = 3.89EE74 pKa = 3.96ISIRR78 pKa = 11.84SLNNSSQVLARR89 pKa = 11.84KK90 pKa = 7.19MARR93 pKa = 11.84TAFNQNDD100 pKa = 3.37KK101 pKa = 10.58SACDD105 pKa = 3.45SGSVSVSISGDD116 pKa = 3.09IADD119 pKa = 5.46DD120 pKa = 3.72EE121 pKa = 4.78TSGNVTAALVASNCRR136 pKa = 11.84SYY138 pKa = 11.3EE139 pKa = 3.86SSDD142 pKa = 3.71GINIEE147 pKa = 3.98GMINGSLSDD156 pKa = 3.69SVSWQTSGSQFTALSSTSTGNLEE179 pKa = 3.51ITYY182 pKa = 10.46VGDD185 pKa = 3.92GSSDD189 pKa = 2.88GHH191 pKa = 6.1NYY193 pKa = 10.42SFGFLYY199 pKa = 10.76DD200 pKa = 4.82LSDD203 pKa = 4.31LNNSMSLNTVNNTLEE218 pKa = 3.62VDD220 pKa = 3.46ANYY223 pKa = 10.84SVYY226 pKa = 10.46IEE228 pKa = 4.57VYY230 pKa = 10.63YY231 pKa = 9.81EE232 pKa = 3.74TDD234 pKa = 3.1NDD236 pKa = 3.74KK237 pKa = 11.24EE238 pKa = 4.21RR239 pKa = 11.84VGGSMKK245 pKa = 10.4VATLDD250 pKa = 3.5SVIYY254 pKa = 9.59DD255 pKa = 3.7APTTTSSLSHH265 pKa = 6.5PVDD268 pKa = 3.06GRR270 pKa = 11.84IQVTAGGQTATVDD283 pKa = 4.67FDD285 pKa = 3.54QTDD288 pKa = 3.72YY289 pKa = 11.18TVTIGNVSNTYY300 pKa = 10.3DD301 pKa = 3.84YY302 pKa = 11.75EE303 pKa = 6.58DD304 pKa = 3.62DD305 pKa = 3.93TVFF308 pKa = 4.52
Molecular weight: 32.69 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0C5VDW0|A0A0C5VDW0_9GAMM Transport permease protein OS=Gynuella sunshinyii YC6258 OX=1445510 GN=YC6258_00346 PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 10.2RR12 pKa = 11.84KK13 pKa = 7.57RR14 pKa = 11.84THH16 pKa = 5.9GFRR19 pKa = 11.84ARR21 pKa = 11.84MATANGRR28 pKa = 11.84QVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.94GRR39 pKa = 11.84KK40 pKa = 8.67VLSAA44 pKa = 4.05
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 10.2RR12 pKa = 11.84KK13 pKa = 7.57RR14 pKa = 11.84THH16 pKa = 5.9GFRR19 pKa = 11.84ARR21 pKa = 11.84MATANGRR28 pKa = 11.84QVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.94GRR39 pKa = 11.84KK40 pKa = 8.67VLSAA44 pKa = 4.05
Molecular weight: 5.11 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1865711 |
37 |
13859 |
315.5 |
35.12 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.577 ± 0.055 | 1.089 ± 0.013 |
5.782 ± 0.046 | 5.838 ± 0.036 |
4.03 ± 0.027 | 7.008 ± 0.045 |
2.447 ± 0.018 | 6.184 ± 0.036 |
4.17 ± 0.049 | 10.397 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.525 ± 0.025 | 4.055 ± 0.032 |
4.173 ± 0.03 | 5.012 ± 0.038 |
5.195 ± 0.027 | 6.706 ± 0.033 |
5.436 ± 0.038 | 6.727 ± 0.027 |
1.447 ± 0.015 | 3.203 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
