
Hubei sobemo-like virus 37
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.64
Get precalculated fractions of proteins
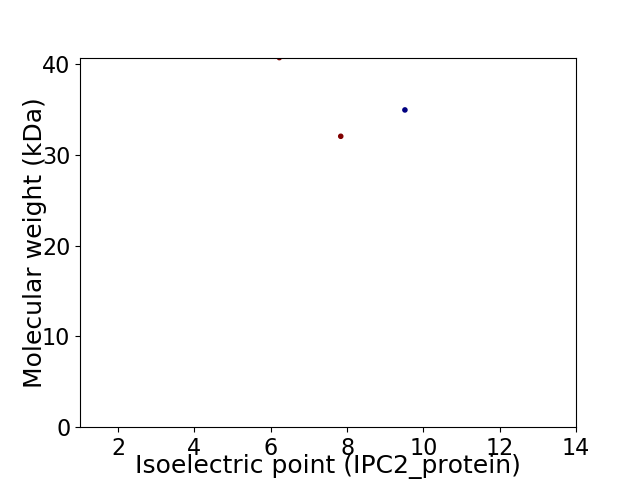
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KEI4|A0A1L3KEI4_9VIRU RNA-directed RNA polymerase OS=Hubei sobemo-like virus 37 OX=1923224 PE=4 SV=1
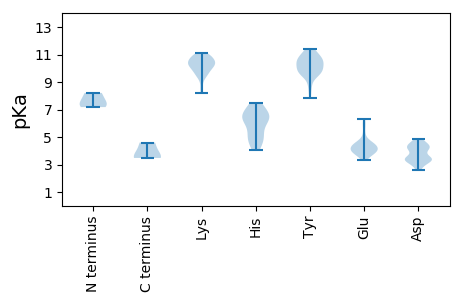
MM1 pKa = 8.21DD2 pKa = 4.34STPGFPFCRR11 pKa = 11.84HH12 pKa = 5.15YY13 pKa = 9.88KK14 pKa = 9.62TNAEE18 pKa = 3.89LLGFNGLNVDD28 pKa = 3.48HH29 pKa = 7.51SKK31 pKa = 8.37TQIVVSSFLKK41 pKa = 9.54WLEE44 pKa = 4.3DD45 pKa = 3.24PVEE48 pKa = 3.99YY49 pKa = 10.42NFRR52 pKa = 11.84MFIKK56 pKa = 10.41DD57 pKa = 3.67EE58 pKa = 3.84PHH60 pKa = 6.51KK61 pKa = 10.21PSKK64 pKa = 10.29VDD66 pKa = 3.13EE67 pKa = 4.55GRR69 pKa = 11.84WRR71 pKa = 11.84LIFSAPLFYY80 pKa = 10.69QILEE84 pKa = 4.22HH85 pKa = 7.18LLLDD89 pKa = 4.69PLDD92 pKa = 4.21EE93 pKa = 5.18LEE95 pKa = 6.28KK96 pKa = 11.01DD97 pKa = 4.19NQWSLPTKK105 pKa = 10.43LGWHH109 pKa = 6.44PFWGGAQMCRR119 pKa = 11.84STFDD123 pKa = 4.14KK124 pKa = 10.24PCSMDD129 pKa = 3.39KK130 pKa = 10.94KK131 pKa = 10.24CWDD134 pKa = 3.29WTLTHH139 pKa = 6.46YY140 pKa = 10.74LVEE143 pKa = 5.67LDD145 pKa = 3.37TQLRR149 pKa = 11.84KK150 pKa = 9.93RR151 pKa = 11.84LVIAPDD157 pKa = 2.57IWYY160 pKa = 9.54DD161 pKa = 3.96LLDD164 pKa = 4.41KK165 pKa = 9.69LTSMSFYY172 pKa = 9.56TASFQFSSGHH182 pKa = 5.3VFRR185 pKa = 11.84QTKK188 pKa = 8.97EE189 pKa = 4.15GIMKK193 pKa = 10.31SGLVRR198 pKa = 11.84TLSTNSHH205 pKa = 5.47CQYY208 pKa = 10.37FIHH211 pKa = 7.27LLACNKK217 pKa = 9.85CGHH220 pKa = 6.01NHH222 pKa = 4.87VCWIIGDD229 pKa = 4.05DD230 pKa = 4.26VIVSDD235 pKa = 4.16PCSEE239 pKa = 3.91YY240 pKa = 11.19LEE242 pKa = 4.5YY243 pKa = 10.35TSHH246 pKa = 6.31YY247 pKa = 10.12CLLKK251 pKa = 10.93DD252 pKa = 4.37VDD254 pKa = 4.08WNYY257 pKa = 10.78HH258 pKa = 4.71FAGFDD263 pKa = 3.5LLKK266 pKa = 10.67NVPLYY271 pKa = 9.6WSKK274 pKa = 10.69HH275 pKa = 4.02VSRR278 pKa = 11.84LLYY281 pKa = 10.82SSDD284 pKa = 3.23EE285 pKa = 4.43VIPEE289 pKa = 3.84ILDD292 pKa = 3.42NYY294 pKa = 8.75QRR296 pKa = 11.84LYY298 pKa = 11.23VYY300 pKa = 10.11DD301 pKa = 3.4QSKK304 pKa = 10.74FNFFQRR310 pKa = 11.84LLAEE314 pKa = 4.27YY315 pKa = 9.87DD316 pKa = 3.47LSKK319 pKa = 10.83LRR321 pKa = 11.84SRR323 pKa = 11.84DD324 pKa = 3.42YY325 pKa = 10.95LLRR328 pKa = 11.84WSVQKK333 pKa = 10.56PSNFKK338 pKa = 10.99FMFKK342 pKa = 9.39TT343 pKa = 3.48
MM1 pKa = 8.21DD2 pKa = 4.34STPGFPFCRR11 pKa = 11.84HH12 pKa = 5.15YY13 pKa = 9.88KK14 pKa = 9.62TNAEE18 pKa = 3.89LLGFNGLNVDD28 pKa = 3.48HH29 pKa = 7.51SKK31 pKa = 8.37TQIVVSSFLKK41 pKa = 9.54WLEE44 pKa = 4.3DD45 pKa = 3.24PVEE48 pKa = 3.99YY49 pKa = 10.42NFRR52 pKa = 11.84MFIKK56 pKa = 10.41DD57 pKa = 3.67EE58 pKa = 3.84PHH60 pKa = 6.51KK61 pKa = 10.21PSKK64 pKa = 10.29VDD66 pKa = 3.13EE67 pKa = 4.55GRR69 pKa = 11.84WRR71 pKa = 11.84LIFSAPLFYY80 pKa = 10.69QILEE84 pKa = 4.22HH85 pKa = 7.18LLLDD89 pKa = 4.69PLDD92 pKa = 4.21EE93 pKa = 5.18LEE95 pKa = 6.28KK96 pKa = 11.01DD97 pKa = 4.19NQWSLPTKK105 pKa = 10.43LGWHH109 pKa = 6.44PFWGGAQMCRR119 pKa = 11.84STFDD123 pKa = 4.14KK124 pKa = 10.24PCSMDD129 pKa = 3.39KK130 pKa = 10.94KK131 pKa = 10.24CWDD134 pKa = 3.29WTLTHH139 pKa = 6.46YY140 pKa = 10.74LVEE143 pKa = 5.67LDD145 pKa = 3.37TQLRR149 pKa = 11.84KK150 pKa = 9.93RR151 pKa = 11.84LVIAPDD157 pKa = 2.57IWYY160 pKa = 9.54DD161 pKa = 3.96LLDD164 pKa = 4.41KK165 pKa = 9.69LTSMSFYY172 pKa = 9.56TASFQFSSGHH182 pKa = 5.3VFRR185 pKa = 11.84QTKK188 pKa = 8.97EE189 pKa = 4.15GIMKK193 pKa = 10.31SGLVRR198 pKa = 11.84TLSTNSHH205 pKa = 5.47CQYY208 pKa = 10.37FIHH211 pKa = 7.27LLACNKK217 pKa = 9.85CGHH220 pKa = 6.01NHH222 pKa = 4.87VCWIIGDD229 pKa = 4.05DD230 pKa = 4.26VIVSDD235 pKa = 4.16PCSEE239 pKa = 3.91YY240 pKa = 11.19LEE242 pKa = 4.5YY243 pKa = 10.35TSHH246 pKa = 6.31YY247 pKa = 10.12CLLKK251 pKa = 10.93DD252 pKa = 4.37VDD254 pKa = 4.08WNYY257 pKa = 10.78HH258 pKa = 4.71FAGFDD263 pKa = 3.5LLKK266 pKa = 10.67NVPLYY271 pKa = 9.6WSKK274 pKa = 10.69HH275 pKa = 4.02VSRR278 pKa = 11.84LLYY281 pKa = 10.82SSDD284 pKa = 3.23EE285 pKa = 4.43VIPEE289 pKa = 3.84ILDD292 pKa = 3.42NYY294 pKa = 8.75QRR296 pKa = 11.84LYY298 pKa = 11.23VYY300 pKa = 10.11DD301 pKa = 3.4QSKK304 pKa = 10.74FNFFQRR310 pKa = 11.84LLAEE314 pKa = 4.27YY315 pKa = 9.87DD316 pKa = 3.47LSKK319 pKa = 10.83LRR321 pKa = 11.84SRR323 pKa = 11.84DD324 pKa = 3.42YY325 pKa = 10.95LLRR328 pKa = 11.84WSVQKK333 pKa = 10.56PSNFKK338 pKa = 10.99FMFKK342 pKa = 9.39TT343 pKa = 3.48
Molecular weight: 40.74 kDa
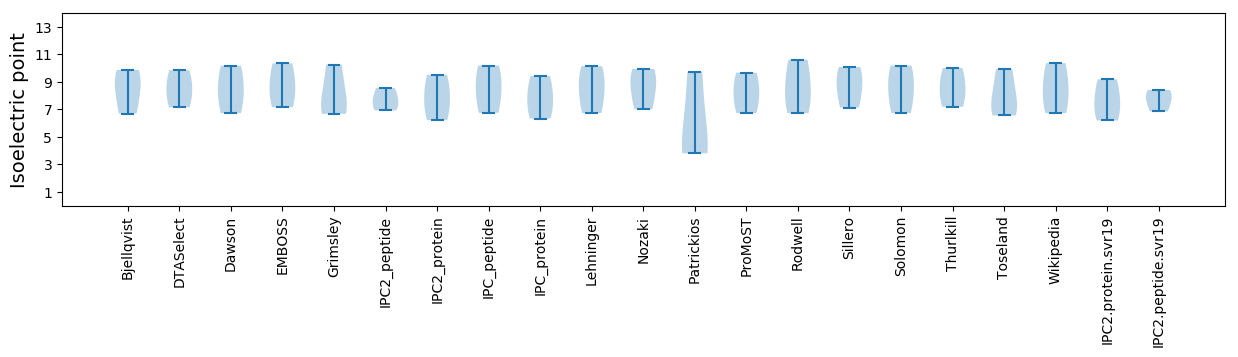
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KEI3|A0A1L3KEI3_9VIRU Uncharacterized protein OS=Hubei sobemo-like virus 37 OX=1923224 PE=4 SV=1
MM1 pKa = 7.18VLNVMLVLGALEE13 pKa = 4.52TILLQHH19 pKa = 6.42HH20 pKa = 6.43MLYY23 pKa = 9.77PVLMVGYY30 pKa = 8.38MFQQLIILLKK40 pKa = 9.36YY41 pKa = 10.08VIIGLYY47 pKa = 9.57IIMMKK52 pKa = 10.16FLSWKK57 pKa = 9.59FLLVFLLLYY66 pKa = 10.76KK67 pKa = 10.39FDD69 pKa = 4.39LLKK72 pKa = 10.73LVPPMIRR79 pKa = 11.84WFVLPDD85 pKa = 3.48VKK87 pKa = 10.74LSNPPLPEE95 pKa = 3.43YY96 pKa = 10.47SYY98 pKa = 10.62PQMYY102 pKa = 10.16LGFIGIVEE110 pKa = 4.02QLSRR114 pKa = 11.84GILEE118 pKa = 4.0LPIWSVIIQLLPCIFLVAILVICVYY143 pKa = 11.16LLVTYY148 pKa = 9.86KK149 pKa = 9.96WLSSSSLSLLDD160 pKa = 4.49LKK162 pKa = 10.48TNLKK166 pKa = 10.98SMVLYY171 pKa = 9.41WLLRR175 pKa = 11.84ILRR178 pKa = 11.84CLVMLVMNLFFRR190 pKa = 11.84GFVLIYY196 pKa = 9.76QGSRR200 pKa = 11.84LLFLKK205 pKa = 10.69AKK207 pKa = 10.43AKK209 pKa = 9.91IKK211 pKa = 10.85RR212 pKa = 11.84EE213 pKa = 4.05DD214 pKa = 2.65VWYY217 pKa = 11.05RR218 pKa = 11.84MIGMKK223 pKa = 10.16KK224 pKa = 9.83IYY226 pKa = 10.28LKK228 pKa = 10.58LKK230 pKa = 10.02KK231 pKa = 10.14YY232 pKa = 9.82VLAVLVLILKK242 pKa = 8.19ITSSNLVNIIILLMMLHH259 pKa = 6.42LVVSKK264 pKa = 10.48SWLVINLFQFILVMLGLMVEE284 pKa = 4.63VSAFLMKK291 pKa = 10.42MMMNLLFF298 pKa = 4.6
MM1 pKa = 7.18VLNVMLVLGALEE13 pKa = 4.52TILLQHH19 pKa = 6.42HH20 pKa = 6.43MLYY23 pKa = 9.77PVLMVGYY30 pKa = 8.38MFQQLIILLKK40 pKa = 9.36YY41 pKa = 10.08VIIGLYY47 pKa = 9.57IIMMKK52 pKa = 10.16FLSWKK57 pKa = 9.59FLLVFLLLYY66 pKa = 10.76KK67 pKa = 10.39FDD69 pKa = 4.39LLKK72 pKa = 10.73LVPPMIRR79 pKa = 11.84WFVLPDD85 pKa = 3.48VKK87 pKa = 10.74LSNPPLPEE95 pKa = 3.43YY96 pKa = 10.47SYY98 pKa = 10.62PQMYY102 pKa = 10.16LGFIGIVEE110 pKa = 4.02QLSRR114 pKa = 11.84GILEE118 pKa = 4.0LPIWSVIIQLLPCIFLVAILVICVYY143 pKa = 11.16LLVTYY148 pKa = 9.86KK149 pKa = 9.96WLSSSSLSLLDD160 pKa = 4.49LKK162 pKa = 10.48TNLKK166 pKa = 10.98SMVLYY171 pKa = 9.41WLLRR175 pKa = 11.84ILRR178 pKa = 11.84CLVMLVMNLFFRR190 pKa = 11.84GFVLIYY196 pKa = 9.76QGSRR200 pKa = 11.84LLFLKK205 pKa = 10.69AKK207 pKa = 10.43AKK209 pKa = 9.91IKK211 pKa = 10.85RR212 pKa = 11.84EE213 pKa = 4.05DD214 pKa = 2.65VWYY217 pKa = 11.05RR218 pKa = 11.84MIGMKK223 pKa = 10.16KK224 pKa = 9.83IYY226 pKa = 10.28LKK228 pKa = 10.58LKK230 pKa = 10.02KK231 pKa = 10.14YY232 pKa = 9.82VLAVLVLILKK242 pKa = 8.19ITSSNLVNIIILLMMLHH259 pKa = 6.42LVVSKK264 pKa = 10.48SWLVINLFQFILVMLGLMVEE284 pKa = 4.63VSAFLMKK291 pKa = 10.42MMMNLLFF298 pKa = 4.6
Molecular weight: 34.97 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
922 |
281 |
343 |
307.3 |
35.93 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
2.82 ± 0.591 | 1.952 ± 0.488 |
5.206 ± 1.719 | 4.23 ± 1.107 |
5.315 ± 0.745 | 4.664 ± 0.993 |
2.386 ± 0.802 | 6.074 ± 1.63 |
6.616 ± 0.368 | 14.751 ± 4.033 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.796 ± 1.584 | 3.688 ± 0.492 |
4.013 ± 0.291 | 3.362 ± 0.319 |
5.098 ± 1.479 | 7.484 ± 0.786 |
3.579 ± 1.004 | 7.592 ± 1.33 |
2.278 ± 0.661 | 5.098 ± 0.068 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
