
Athene cunicularia (Burrowing owl) (Speotyto cunicularia)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Deuterostomia; Chordata; Craniata; Vertebrata; Gnathostomata; Teleostomi; Euteleostomi; Sarcopterygii; Dipnotetrapodomorpha; Tetrapoda; Amniota; Sauropsida; Sauria; Archelosauria; Archosauria;
Average proteome isoelectric point is 6.56
Get precalculated fractions of proteins
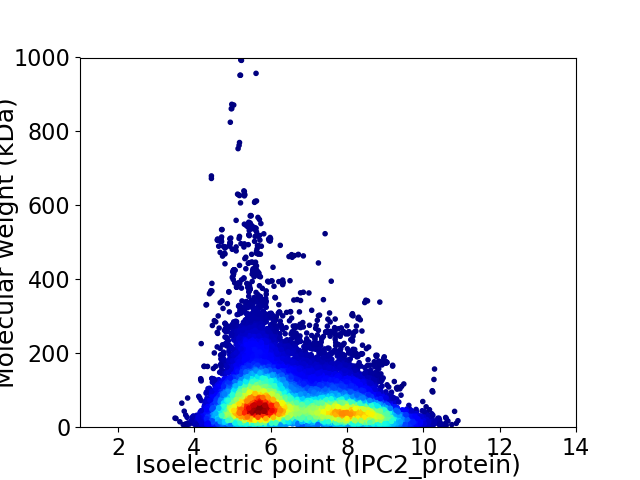
Virtual 2D-PAGE plot for 22627 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A663N5P3|A0A663N5P3_ATHCN Myosin heavy chain 11 OS=Athene cunicularia OX=194338 GN=MYH11 PE=3 SV=1
MM1 pKa = 6.93GTEE4 pKa = 4.24SLSPEE9 pKa = 4.26GPALVPRR16 pKa = 11.84GYY18 pKa = 11.25NKK20 pKa = 10.12LHH22 pKa = 6.27SVNLQHH28 pKa = 6.94AHH30 pKa = 5.27VADD33 pKa = 4.47GQSHH37 pKa = 6.22SIIVRR42 pKa = 11.84LSGLRR47 pKa = 11.84GDD49 pKa = 3.9MLSVEE54 pKa = 5.06LYY56 pKa = 10.52VDD58 pKa = 4.55CKK60 pKa = 11.21QMDD63 pKa = 3.79SSVGLPEE70 pKa = 4.7LSEE73 pKa = 4.13IPLAEE78 pKa = 4.06VEE80 pKa = 4.5SIEE83 pKa = 4.74VRR85 pKa = 11.84TGQKK89 pKa = 10.77GFVEE93 pKa = 4.33SMKK96 pKa = 10.78LILGGSMSRR105 pKa = 11.84VGALSEE111 pKa = 4.81CPFQGDD117 pKa = 3.71EE118 pKa = 4.7SIHH121 pKa = 5.77SAGNGTAAPCTEE133 pKa = 3.67QGQRR137 pKa = 11.84GGLTGSPVHH146 pKa = 5.57ATARR150 pKa = 11.84SAVSAGEE157 pKa = 3.98APLPPRR163 pKa = 11.84GAKK166 pKa = 9.22GRR168 pKa = 11.84PASPPPWPSLGFHH181 pKa = 6.0EE182 pKa = 5.12HH183 pKa = 7.6RR184 pKa = 11.84SRR186 pKa = 11.84CNPNPCFSGVDD197 pKa = 3.37CMEE200 pKa = 4.13TYY202 pKa = 10.02EE203 pKa = 4.17YY204 pKa = 10.08PGYY207 pKa = 10.36RR208 pKa = 11.84CGPCPPGLEE217 pKa = 4.47GNGTSCADD225 pKa = 3.4INEE228 pKa = 4.28CAYY231 pKa = 10.69ANPCFPGSKK240 pKa = 9.86CINTAPGFRR249 pKa = 11.84CEE251 pKa = 4.01PCPRR255 pKa = 11.84GYY257 pKa = 10.53RR258 pKa = 11.84GNTVSGVGVDD268 pKa = 3.41YY269 pKa = 11.37AKK271 pKa = 10.72ASKK274 pKa = 9.68QVCTDD279 pKa = 2.78IDD281 pKa = 3.7EE282 pKa = 5.35CNDD285 pKa = 3.58GNNGGCDD292 pKa = 3.53PNSICTNTLGSYY304 pKa = 10.15KK305 pKa = 10.38CGPCKK310 pKa = 10.49SGFVGNQTSGCIPQKK325 pKa = 10.52SCSTPTSNPCDD336 pKa = 3.41INGFCMFEE344 pKa = 4.27RR345 pKa = 11.84NGEE348 pKa = 3.95ISCACNVGWAGNGNVCGQDD367 pKa = 3.15TDD369 pKa = 4.23LDD371 pKa = 4.44GYY373 pKa = 9.92PDD375 pKa = 4.1EE376 pKa = 5.31PLPCIDD382 pKa = 4.25NNKK385 pKa = 9.35HH386 pKa = 6.21CKK388 pKa = 9.14QDD390 pKa = 3.44NCRR393 pKa = 11.84LTPNSGQEE401 pKa = 3.95DD402 pKa = 4.26ADD404 pKa = 3.62NDD406 pKa = 5.11GIGDD410 pKa = 3.82QCDD413 pKa = 3.57DD414 pKa = 4.52DD415 pKa = 6.87ADD417 pKa = 4.09GDD419 pKa = 4.3GIKK422 pKa = 10.54NVEE425 pKa = 3.95VDD427 pKa = 3.34NCRR430 pKa = 11.84LFPNKK435 pKa = 9.91DD436 pKa = 3.46QQNSDD441 pKa = 3.11TDD443 pKa = 4.03SFGDD447 pKa = 4.2ACDD450 pKa = 3.62NCPNVPNNDD459 pKa = 3.4QRR461 pKa = 11.84DD462 pKa = 3.62TDD464 pKa = 4.31SNGEE468 pKa = 3.91GDD470 pKa = 4.42ACDD473 pKa = 3.98NDD475 pKa = 3.39IDD477 pKa = 5.15GDD479 pKa = 4.46GIPNMLDD486 pKa = 2.96NCPRR490 pKa = 11.84VPNPLQTDD498 pKa = 3.4RR499 pKa = 11.84DD500 pKa = 3.67EE501 pKa = 6.42DD502 pKa = 4.4GVGDD506 pKa = 4.27ACDD509 pKa = 3.66SCPEE513 pKa = 4.12MSNPTQTDD521 pKa = 3.06MDD523 pKa = 4.69SDD525 pKa = 4.47LVGDD529 pKa = 3.68ICDD532 pKa = 3.85TNEE535 pKa = 4.14DD536 pKa = 3.74SDD538 pKa = 4.93GDD540 pKa = 3.91GHH542 pKa = 7.38QDD544 pKa = 3.18TKK546 pKa = 11.57DD547 pKa = 3.32NCAEE551 pKa = 4.25IPNSSQLDD559 pKa = 3.58SDD561 pKa = 4.01NDD563 pKa = 4.11GLGDD567 pKa = 4.98DD568 pKa = 5.48CDD570 pKa = 4.94NDD572 pKa = 4.37DD573 pKa = 5.42DD574 pKa = 5.52NDD576 pKa = 5.09GIPDD580 pKa = 4.02YY581 pKa = 11.2VPPGPDD587 pKa = 2.98NCRR590 pKa = 11.84LIPNPNQKK598 pKa = 10.66DD599 pKa = 3.36SDD601 pKa = 4.17GNGVGDD607 pKa = 3.6VCEE610 pKa = 4.36EE611 pKa = 4.28DD612 pKa = 3.68FDD614 pKa = 5.16NDD616 pKa = 4.06TVVDD620 pKa = 3.93QLDD623 pKa = 3.73VCPEE627 pKa = 3.98SAEE630 pKa = 4.05VTLTDD635 pKa = 3.82FRR637 pKa = 11.84AYY639 pKa = 8.36QTVILDD645 pKa = 3.91PEE647 pKa = 5.2GDD649 pKa = 3.69AQIDD653 pKa = 4.13PNWVVLNQGMEE664 pKa = 4.11IVQTMNSDD672 pKa = 3.19PGLAVGYY679 pKa = 7.31TAFNGVDD686 pKa = 3.99FEE688 pKa = 4.97GTFHH692 pKa = 6.81VNTVTDD698 pKa = 3.57DD699 pKa = 4.08DD700 pKa = 4.34YY701 pKa = 12.06AGFIFSYY708 pKa = 10.41QDD710 pKa = 2.92SASFYY715 pKa = 10.36VVMWKK720 pKa = 7.78QTEE723 pKa = 3.86QTYY726 pKa = 8.13WQATPFRR733 pKa = 11.84AVAEE737 pKa = 4.58PGLQQ741 pKa = 3.16
MM1 pKa = 6.93GTEE4 pKa = 4.24SLSPEE9 pKa = 4.26GPALVPRR16 pKa = 11.84GYY18 pKa = 11.25NKK20 pKa = 10.12LHH22 pKa = 6.27SVNLQHH28 pKa = 6.94AHH30 pKa = 5.27VADD33 pKa = 4.47GQSHH37 pKa = 6.22SIIVRR42 pKa = 11.84LSGLRR47 pKa = 11.84GDD49 pKa = 3.9MLSVEE54 pKa = 5.06LYY56 pKa = 10.52VDD58 pKa = 4.55CKK60 pKa = 11.21QMDD63 pKa = 3.79SSVGLPEE70 pKa = 4.7LSEE73 pKa = 4.13IPLAEE78 pKa = 4.06VEE80 pKa = 4.5SIEE83 pKa = 4.74VRR85 pKa = 11.84TGQKK89 pKa = 10.77GFVEE93 pKa = 4.33SMKK96 pKa = 10.78LILGGSMSRR105 pKa = 11.84VGALSEE111 pKa = 4.81CPFQGDD117 pKa = 3.71EE118 pKa = 4.7SIHH121 pKa = 5.77SAGNGTAAPCTEE133 pKa = 3.67QGQRR137 pKa = 11.84GGLTGSPVHH146 pKa = 5.57ATARR150 pKa = 11.84SAVSAGEE157 pKa = 3.98APLPPRR163 pKa = 11.84GAKK166 pKa = 9.22GRR168 pKa = 11.84PASPPPWPSLGFHH181 pKa = 6.0EE182 pKa = 5.12HH183 pKa = 7.6RR184 pKa = 11.84SRR186 pKa = 11.84CNPNPCFSGVDD197 pKa = 3.37CMEE200 pKa = 4.13TYY202 pKa = 10.02EE203 pKa = 4.17YY204 pKa = 10.08PGYY207 pKa = 10.36RR208 pKa = 11.84CGPCPPGLEE217 pKa = 4.47GNGTSCADD225 pKa = 3.4INEE228 pKa = 4.28CAYY231 pKa = 10.69ANPCFPGSKK240 pKa = 9.86CINTAPGFRR249 pKa = 11.84CEE251 pKa = 4.01PCPRR255 pKa = 11.84GYY257 pKa = 10.53RR258 pKa = 11.84GNTVSGVGVDD268 pKa = 3.41YY269 pKa = 11.37AKK271 pKa = 10.72ASKK274 pKa = 9.68QVCTDD279 pKa = 2.78IDD281 pKa = 3.7EE282 pKa = 5.35CNDD285 pKa = 3.58GNNGGCDD292 pKa = 3.53PNSICTNTLGSYY304 pKa = 10.15KK305 pKa = 10.38CGPCKK310 pKa = 10.49SGFVGNQTSGCIPQKK325 pKa = 10.52SCSTPTSNPCDD336 pKa = 3.41INGFCMFEE344 pKa = 4.27RR345 pKa = 11.84NGEE348 pKa = 3.95ISCACNVGWAGNGNVCGQDD367 pKa = 3.15TDD369 pKa = 4.23LDD371 pKa = 4.44GYY373 pKa = 9.92PDD375 pKa = 4.1EE376 pKa = 5.31PLPCIDD382 pKa = 4.25NNKK385 pKa = 9.35HH386 pKa = 6.21CKK388 pKa = 9.14QDD390 pKa = 3.44NCRR393 pKa = 11.84LTPNSGQEE401 pKa = 3.95DD402 pKa = 4.26ADD404 pKa = 3.62NDD406 pKa = 5.11GIGDD410 pKa = 3.82QCDD413 pKa = 3.57DD414 pKa = 4.52DD415 pKa = 6.87ADD417 pKa = 4.09GDD419 pKa = 4.3GIKK422 pKa = 10.54NVEE425 pKa = 3.95VDD427 pKa = 3.34NCRR430 pKa = 11.84LFPNKK435 pKa = 9.91DD436 pKa = 3.46QQNSDD441 pKa = 3.11TDD443 pKa = 4.03SFGDD447 pKa = 4.2ACDD450 pKa = 3.62NCPNVPNNDD459 pKa = 3.4QRR461 pKa = 11.84DD462 pKa = 3.62TDD464 pKa = 4.31SNGEE468 pKa = 3.91GDD470 pKa = 4.42ACDD473 pKa = 3.98NDD475 pKa = 3.39IDD477 pKa = 5.15GDD479 pKa = 4.46GIPNMLDD486 pKa = 2.96NCPRR490 pKa = 11.84VPNPLQTDD498 pKa = 3.4RR499 pKa = 11.84DD500 pKa = 3.67EE501 pKa = 6.42DD502 pKa = 4.4GVGDD506 pKa = 4.27ACDD509 pKa = 3.66SCPEE513 pKa = 4.12MSNPTQTDD521 pKa = 3.06MDD523 pKa = 4.69SDD525 pKa = 4.47LVGDD529 pKa = 3.68ICDD532 pKa = 3.85TNEE535 pKa = 4.14DD536 pKa = 3.74SDD538 pKa = 4.93GDD540 pKa = 3.91GHH542 pKa = 7.38QDD544 pKa = 3.18TKK546 pKa = 11.57DD547 pKa = 3.32NCAEE551 pKa = 4.25IPNSSQLDD559 pKa = 3.58SDD561 pKa = 4.01NDD563 pKa = 4.11GLGDD567 pKa = 4.98DD568 pKa = 5.48CDD570 pKa = 4.94NDD572 pKa = 4.37DD573 pKa = 5.42DD574 pKa = 5.52NDD576 pKa = 5.09GIPDD580 pKa = 4.02YY581 pKa = 11.2VPPGPDD587 pKa = 2.98NCRR590 pKa = 11.84LIPNPNQKK598 pKa = 10.66DD599 pKa = 3.36SDD601 pKa = 4.17GNGVGDD607 pKa = 3.6VCEE610 pKa = 4.36EE611 pKa = 4.28DD612 pKa = 3.68FDD614 pKa = 5.16NDD616 pKa = 4.06TVVDD620 pKa = 3.93QLDD623 pKa = 3.73VCPEE627 pKa = 3.98SAEE630 pKa = 4.05VTLTDD635 pKa = 3.82FRR637 pKa = 11.84AYY639 pKa = 8.36QTVILDD645 pKa = 3.91PEE647 pKa = 5.2GDD649 pKa = 3.69AQIDD653 pKa = 4.13PNWVVLNQGMEE664 pKa = 4.11IVQTMNSDD672 pKa = 3.19PGLAVGYY679 pKa = 7.31TAFNGVDD686 pKa = 3.99FEE688 pKa = 4.97GTFHH692 pKa = 6.81VNTVTDD698 pKa = 3.57DD699 pKa = 4.08DD700 pKa = 4.34YY701 pKa = 12.06AGFIFSYY708 pKa = 10.41QDD710 pKa = 2.92SASFYY715 pKa = 10.36VVMWKK720 pKa = 7.78QTEE723 pKa = 3.86QTYY726 pKa = 8.13WQATPFRR733 pKa = 11.84AVAEE737 pKa = 4.58PGLQQ741 pKa = 3.16
Molecular weight: 78.93 kDa
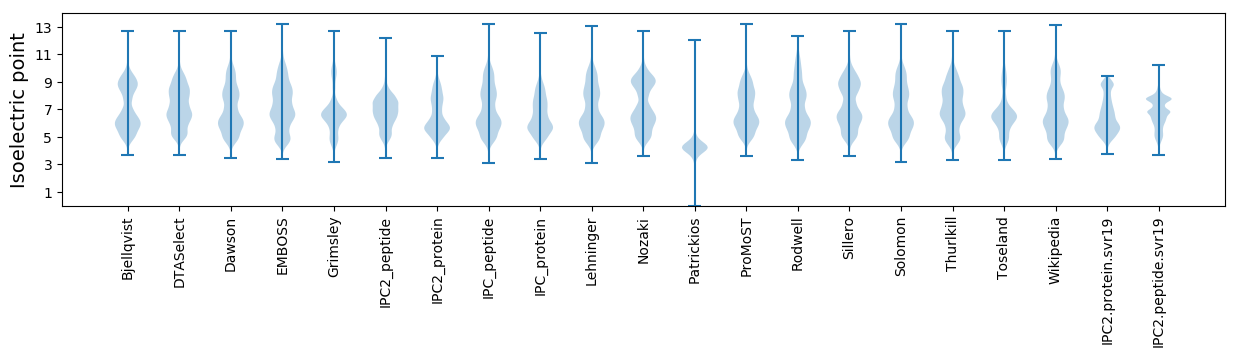
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A663M9F8|A0A663M9F8_ATHCN T-complex protein 1 subunit gamma OS=Athene cunicularia OX=194338 PE=3 SV=1
II1 pKa = 7.08PRR3 pKa = 11.84PRR5 pKa = 11.84GAGSPRR11 pKa = 11.84PRR13 pKa = 11.84GAGSPRR19 pKa = 11.84PRR21 pKa = 11.84GAGSPRR27 pKa = 11.84PRR29 pKa = 11.84GAGSPRR35 pKa = 11.84PRR37 pKa = 11.84GAGSPRR43 pKa = 11.84PRR45 pKa = 11.84GAGSPRR51 pKa = 11.84PRR53 pKa = 11.84GAGSPRR59 pKa = 11.84PRR61 pKa = 11.84GAGSPRR67 pKa = 11.84PRR69 pKa = 11.84GAGSPRR75 pKa = 11.84PRR77 pKa = 11.84GAGSPRR83 pKa = 11.84PRR85 pKa = 11.84GAGSPRR91 pKa = 11.84PRR93 pKa = 11.84GAGSPRR99 pKa = 11.84PRR101 pKa = 11.84GAGSPRR107 pKa = 11.84PRR109 pKa = 11.84GAGSPWPRR117 pKa = 11.84GAGSPWPRR125 pKa = 11.84GAGSVTSRR133 pKa = 11.84APRR136 pKa = 11.84GTDD139 pKa = 2.94HH140 pKa = 7.27AGASAGEE147 pKa = 3.89RR148 pKa = 11.84EE149 pKa = 4.85GWRR152 pKa = 11.84EE153 pKa = 3.84HH154 pKa = 6.56PSWKK158 pKa = 10.09AAAPLHH164 pKa = 5.97SVLPAGPRR172 pKa = 11.84PAPCC176 pKa = 4.62
II1 pKa = 7.08PRR3 pKa = 11.84PRR5 pKa = 11.84GAGSPRR11 pKa = 11.84PRR13 pKa = 11.84GAGSPRR19 pKa = 11.84PRR21 pKa = 11.84GAGSPRR27 pKa = 11.84PRR29 pKa = 11.84GAGSPRR35 pKa = 11.84PRR37 pKa = 11.84GAGSPRR43 pKa = 11.84PRR45 pKa = 11.84GAGSPRR51 pKa = 11.84PRR53 pKa = 11.84GAGSPRR59 pKa = 11.84PRR61 pKa = 11.84GAGSPRR67 pKa = 11.84PRR69 pKa = 11.84GAGSPRR75 pKa = 11.84PRR77 pKa = 11.84GAGSPRR83 pKa = 11.84PRR85 pKa = 11.84GAGSPRR91 pKa = 11.84PRR93 pKa = 11.84GAGSPRR99 pKa = 11.84PRR101 pKa = 11.84GAGSPRR107 pKa = 11.84PRR109 pKa = 11.84GAGSPWPRR117 pKa = 11.84GAGSPWPRR125 pKa = 11.84GAGSVTSRR133 pKa = 11.84APRR136 pKa = 11.84GTDD139 pKa = 2.94HH140 pKa = 7.27AGASAGEE147 pKa = 3.89RR148 pKa = 11.84EE149 pKa = 4.85GWRR152 pKa = 11.84EE153 pKa = 3.84HH154 pKa = 6.56PSWKK158 pKa = 10.09AAAPLHH164 pKa = 5.97SVLPAGPRR172 pKa = 11.84PAPCC176 pKa = 4.62
Molecular weight: 17.53 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
15598483 |
16 |
8698 |
689.4 |
77.26 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.412 ± 0.011 | 2.281 ± 0.013 |
5.053 ± 0.011 | 7.21 ± 0.023 |
3.82 ± 0.012 | 5.944 ± 0.018 |
2.536 ± 0.007 | 4.937 ± 0.012 |
6.239 ± 0.015 | 9.729 ± 0.018 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.255 ± 0.006 | 4.131 ± 0.01 |
5.357 ± 0.019 | 4.746 ± 0.016 |
5.18 ± 0.011 | 8.329 ± 0.019 |
5.45 ± 0.01 | 6.235 ± 0.011 |
1.193 ± 0.005 | 2.889 ± 0.009 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
