
Rice grassy stunt tenuivirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Polyploviricotina; Ellioviricetes; Bunyavirales; Phenuiviridae; Tenuivirus
Average proteome isoelectric point is 6.41
Get precalculated fractions of proteins
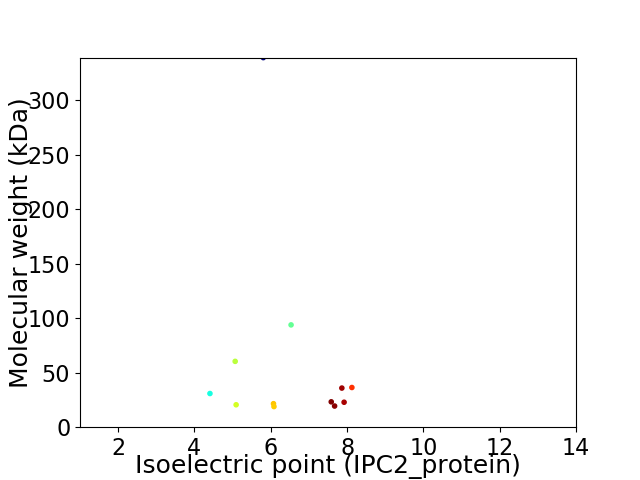
Virtual 2D-PAGE plot for 12 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|O89174|O89174_9VIRU p4V protein OS=Rice grassy stunt tenuivirus OX=66266 GN=p4V PE=4 SV=1
MM1 pKa = 7.48FSLSDD6 pKa = 3.64FLSEE10 pKa = 3.53IGNVIRR16 pKa = 11.84EE17 pKa = 4.26VEE19 pKa = 4.09DD20 pKa = 3.61KK21 pKa = 11.13DD22 pKa = 3.54NKK24 pKa = 10.46EE25 pKa = 4.0YY26 pKa = 10.71EE27 pKa = 4.06DD28 pKa = 4.33QIRR31 pKa = 11.84EE32 pKa = 4.04IQNLHH37 pKa = 6.47RR38 pKa = 11.84DD39 pKa = 3.69MNHH42 pKa = 5.32VEE44 pKa = 4.39LKK46 pKa = 10.07VSEE49 pKa = 4.59NIKK52 pKa = 10.08KK53 pKa = 10.23ACPEE57 pKa = 3.95LTEE60 pKa = 4.34TEE62 pKa = 4.5ANLSSDD68 pKa = 3.31NTIIDD73 pKa = 4.28LKK75 pKa = 11.2DD76 pKa = 3.6DD77 pKa = 4.05ALTEE81 pKa = 4.1YY82 pKa = 11.01KK83 pKa = 10.55EE84 pKa = 4.25IVWKK88 pKa = 10.25IDD90 pKa = 3.1NDD92 pKa = 3.65EE93 pKa = 4.1SVEE96 pKa = 4.03MSKK99 pKa = 8.57NTIKK103 pKa = 10.92EE104 pKa = 4.07MVFKK108 pKa = 11.23NKK110 pKa = 9.85DD111 pKa = 3.3DD112 pKa = 3.87EE113 pKa = 5.52LIISSNLDD121 pKa = 3.05LDD123 pKa = 4.31QNVCNGNSDD132 pKa = 4.06CDD134 pKa = 3.58VSNAKK139 pKa = 10.24KK140 pKa = 10.63SEE142 pKa = 3.86NGEE145 pKa = 4.17NFLEE149 pKa = 4.48THH151 pKa = 6.5TDD153 pKa = 3.83MIPPINTSDD162 pKa = 3.65CNHH165 pKa = 6.42INEE168 pKa = 4.48ILDD171 pKa = 3.46NGTYY175 pKa = 10.03EE176 pKa = 4.66RR177 pKa = 11.84IAIFSNPITINLDD190 pKa = 3.43DD191 pKa = 4.05GRR193 pKa = 11.84KK194 pKa = 9.28LIKK197 pKa = 9.64VTIADD202 pKa = 3.75EE203 pKa = 4.49NIKK206 pKa = 10.68KK207 pKa = 10.31KK208 pKa = 10.92VEE210 pKa = 4.14SSVKK214 pKa = 10.23EE215 pKa = 3.82ITCIPIYY222 pKa = 10.59KK223 pKa = 9.9LGNRR227 pKa = 11.84GDD229 pKa = 3.76YY230 pKa = 11.01DD231 pKa = 5.03SIISNCMSMDD241 pKa = 3.48GVIKK245 pKa = 10.73KK246 pKa = 8.3LTGTLIIYY254 pKa = 10.07CSLDD258 pKa = 4.11DD259 pKa = 3.84VFSWLILKK267 pKa = 8.55SVPNDD272 pKa = 3.14
MM1 pKa = 7.48FSLSDD6 pKa = 3.64FLSEE10 pKa = 3.53IGNVIRR16 pKa = 11.84EE17 pKa = 4.26VEE19 pKa = 4.09DD20 pKa = 3.61KK21 pKa = 11.13DD22 pKa = 3.54NKK24 pKa = 10.46EE25 pKa = 4.0YY26 pKa = 10.71EE27 pKa = 4.06DD28 pKa = 4.33QIRR31 pKa = 11.84EE32 pKa = 4.04IQNLHH37 pKa = 6.47RR38 pKa = 11.84DD39 pKa = 3.69MNHH42 pKa = 5.32VEE44 pKa = 4.39LKK46 pKa = 10.07VSEE49 pKa = 4.59NIKK52 pKa = 10.08KK53 pKa = 10.23ACPEE57 pKa = 3.95LTEE60 pKa = 4.34TEE62 pKa = 4.5ANLSSDD68 pKa = 3.31NTIIDD73 pKa = 4.28LKK75 pKa = 11.2DD76 pKa = 3.6DD77 pKa = 4.05ALTEE81 pKa = 4.1YY82 pKa = 11.01KK83 pKa = 10.55EE84 pKa = 4.25IVWKK88 pKa = 10.25IDD90 pKa = 3.1NDD92 pKa = 3.65EE93 pKa = 4.1SVEE96 pKa = 4.03MSKK99 pKa = 8.57NTIKK103 pKa = 10.92EE104 pKa = 4.07MVFKK108 pKa = 11.23NKK110 pKa = 9.85DD111 pKa = 3.3DD112 pKa = 3.87EE113 pKa = 5.52LIISSNLDD121 pKa = 3.05LDD123 pKa = 4.31QNVCNGNSDD132 pKa = 4.06CDD134 pKa = 3.58VSNAKK139 pKa = 10.24KK140 pKa = 10.63SEE142 pKa = 3.86NGEE145 pKa = 4.17NFLEE149 pKa = 4.48THH151 pKa = 6.5TDD153 pKa = 3.83MIPPINTSDD162 pKa = 3.65CNHH165 pKa = 6.42INEE168 pKa = 4.48ILDD171 pKa = 3.46NGTYY175 pKa = 10.03EE176 pKa = 4.66RR177 pKa = 11.84IAIFSNPITINLDD190 pKa = 3.43DD191 pKa = 4.05GRR193 pKa = 11.84KK194 pKa = 9.28LIKK197 pKa = 9.64VTIADD202 pKa = 3.75EE203 pKa = 4.49NIKK206 pKa = 10.68KK207 pKa = 10.31KK208 pKa = 10.92VEE210 pKa = 4.14SSVKK214 pKa = 10.23EE215 pKa = 3.82ITCIPIYY222 pKa = 10.59KK223 pKa = 9.9LGNRR227 pKa = 11.84GDD229 pKa = 3.76YY230 pKa = 11.01DD231 pKa = 5.03SIISNCMSMDD241 pKa = 3.48GVIKK245 pKa = 10.73KK246 pKa = 8.3LTGTLIIYY254 pKa = 10.07CSLDD258 pKa = 4.11DD259 pKa = 3.84VFSWLILKK267 pKa = 8.55SVPNDD272 pKa = 3.14
Molecular weight: 30.91 kDa
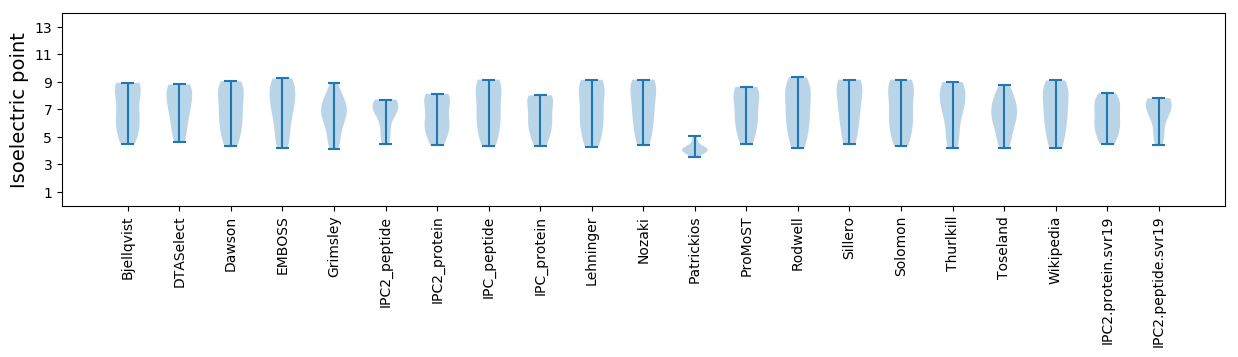
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|O89162|O89162_9VIRU 18.9K protein OS=Rice grassy stunt tenuivirus OX=66266 GN=pV1 PE=4 SV=1
MM1 pKa = 7.89ALLQKK6 pKa = 10.59LGSSKK11 pKa = 10.45VSSKK15 pKa = 10.69RR16 pKa = 11.84MSPAMIPLDD25 pKa = 5.03SINQDD30 pKa = 3.14LVDD33 pKa = 4.12PQQEE37 pKa = 3.68KK38 pKa = 10.33DD39 pKa = 3.35AKK41 pKa = 10.28NKK43 pKa = 10.45KK44 pKa = 8.77EE45 pKa = 4.11GKK47 pKa = 10.16KK48 pKa = 9.96KK49 pKa = 10.7DD50 pKa = 4.13LDD52 pKa = 3.65VSMDD56 pKa = 3.94PLTGKK61 pKa = 10.57LPLGKK66 pKa = 9.91KK67 pKa = 9.49KK68 pKa = 10.4QVDD71 pKa = 3.58TGGIAYY77 pKa = 10.21LEE79 pKa = 4.23NALMQLDD86 pKa = 3.83LHH88 pKa = 6.91DD89 pKa = 4.87FSFDD93 pKa = 3.7SIRR96 pKa = 11.84PRR98 pKa = 11.84TKK100 pKa = 8.32TFHH103 pKa = 6.77MKK105 pKa = 9.86RR106 pKa = 11.84QHH108 pKa = 6.16FKK110 pKa = 10.33ISTVNSRR117 pKa = 11.84FRR119 pKa = 11.84LDD121 pKa = 3.38VEE123 pKa = 4.29KK124 pKa = 10.27TGLFSKK130 pKa = 8.16TLKK133 pKa = 10.28YY134 pKa = 10.96SRR136 pKa = 11.84ICTLCLAFLGIKK148 pKa = 9.83NRR150 pKa = 11.84AQGTISFTFRR160 pKa = 11.84DD161 pKa = 3.68LSYY164 pKa = 11.02LSEE167 pKa = 4.15NDD169 pKa = 3.47QIDD172 pKa = 4.08FKK174 pKa = 11.44VKK176 pKa = 10.34NRR178 pKa = 11.84ISKK181 pKa = 9.99SFSAIASFPAPIFNDD196 pKa = 3.3DD197 pKa = 4.59LGNLICDD204 pKa = 4.22FEE206 pKa = 4.69IEE208 pKa = 4.21NASVNGVVIGDD219 pKa = 4.06LLVLLGIEE227 pKa = 4.21QSDD230 pKa = 4.08LPVCYY235 pKa = 9.66EE236 pKa = 3.8PQKK239 pKa = 11.46AKK241 pKa = 10.38IFEE244 pKa = 4.32YY245 pKa = 10.78KK246 pKa = 10.35PLTEE250 pKa = 4.7KK251 pKa = 10.96GLNKK255 pKa = 9.87ISNFAGYY262 pKa = 10.15VDD264 pKa = 4.49NVLKK268 pKa = 10.73AAINHH273 pKa = 6.57RR274 pKa = 11.84EE275 pKa = 4.04GEE277 pKa = 4.07DD278 pKa = 3.42DD279 pKa = 3.97GFSTEE284 pKa = 3.92GLGVLVHH291 pKa = 6.14PRR293 pKa = 11.84VKK295 pKa = 10.64QIDD298 pKa = 3.38NSIPIKK304 pKa = 10.61SLEE307 pKa = 4.07NKK309 pKa = 7.42PQKK312 pKa = 10.33MLMRR316 pKa = 11.84DD317 pKa = 3.43GSYY320 pKa = 11.28LDD322 pKa = 3.81VNPP325 pKa = 4.65
MM1 pKa = 7.89ALLQKK6 pKa = 10.59LGSSKK11 pKa = 10.45VSSKK15 pKa = 10.69RR16 pKa = 11.84MSPAMIPLDD25 pKa = 5.03SINQDD30 pKa = 3.14LVDD33 pKa = 4.12PQQEE37 pKa = 3.68KK38 pKa = 10.33DD39 pKa = 3.35AKK41 pKa = 10.28NKK43 pKa = 10.45KK44 pKa = 8.77EE45 pKa = 4.11GKK47 pKa = 10.16KK48 pKa = 9.96KK49 pKa = 10.7DD50 pKa = 4.13LDD52 pKa = 3.65VSMDD56 pKa = 3.94PLTGKK61 pKa = 10.57LPLGKK66 pKa = 9.91KK67 pKa = 9.49KK68 pKa = 10.4QVDD71 pKa = 3.58TGGIAYY77 pKa = 10.21LEE79 pKa = 4.23NALMQLDD86 pKa = 3.83LHH88 pKa = 6.91DD89 pKa = 4.87FSFDD93 pKa = 3.7SIRR96 pKa = 11.84PRR98 pKa = 11.84TKK100 pKa = 8.32TFHH103 pKa = 6.77MKK105 pKa = 9.86RR106 pKa = 11.84QHH108 pKa = 6.16FKK110 pKa = 10.33ISTVNSRR117 pKa = 11.84FRR119 pKa = 11.84LDD121 pKa = 3.38VEE123 pKa = 4.29KK124 pKa = 10.27TGLFSKK130 pKa = 8.16TLKK133 pKa = 10.28YY134 pKa = 10.96SRR136 pKa = 11.84ICTLCLAFLGIKK148 pKa = 9.83NRR150 pKa = 11.84AQGTISFTFRR160 pKa = 11.84DD161 pKa = 3.68LSYY164 pKa = 11.02LSEE167 pKa = 4.15NDD169 pKa = 3.47QIDD172 pKa = 4.08FKK174 pKa = 11.44VKK176 pKa = 10.34NRR178 pKa = 11.84ISKK181 pKa = 9.99SFSAIASFPAPIFNDD196 pKa = 3.3DD197 pKa = 4.59LGNLICDD204 pKa = 4.22FEE206 pKa = 4.69IEE208 pKa = 4.21NASVNGVVIGDD219 pKa = 4.06LLVLLGIEE227 pKa = 4.21QSDD230 pKa = 4.08LPVCYY235 pKa = 9.66EE236 pKa = 3.8PQKK239 pKa = 11.46AKK241 pKa = 10.38IFEE244 pKa = 4.32YY245 pKa = 10.78KK246 pKa = 10.35PLTEE250 pKa = 4.7KK251 pKa = 10.96GLNKK255 pKa = 9.87ISNFAGYY262 pKa = 10.15VDD264 pKa = 4.49NVLKK268 pKa = 10.73AAINHH273 pKa = 6.57RR274 pKa = 11.84EE275 pKa = 4.04GEE277 pKa = 4.07DD278 pKa = 3.42DD279 pKa = 3.97GFSTEE284 pKa = 3.92GLGVLVHH291 pKa = 6.14PRR293 pKa = 11.84VKK295 pKa = 10.64QIDD298 pKa = 3.38NSIPIKK304 pKa = 10.61SLEE307 pKa = 4.07NKK309 pKa = 7.42PQKK312 pKa = 10.33MLMRR316 pKa = 11.84DD317 pKa = 3.43GSYY320 pKa = 11.28LDD322 pKa = 3.81VNPP325 pKa = 4.65
Molecular weight: 36.42 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
6290 |
165 |
2925 |
524.2 |
60.28 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.529 ± 0.515 | 2.003 ± 0.331 |
6.979 ± 0.438 | 6.439 ± 0.517 |
4.165 ± 0.296 | 4.785 ± 0.254 |
2.226 ± 0.129 | 7.107 ± 0.622 |
7.393 ± 0.353 | 9.905 ± 0.324 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.275 ± 0.328 | 5.517 ± 0.443 |
3.084 ± 0.217 | 3.18 ± 0.28 |
4.992 ± 0.291 | 8.378 ± 0.378 |
5.501 ± 0.387 | 6.216 ± 0.512 |
1.065 ± 0.095 | 4.261 ± 0.287 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
