
Chicken associated smacovirus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cremevirales; Smacoviridae; Huchismacovirus; unclassified Huchismacovirus
Average proteome isoelectric point is 6.94
Get precalculated fractions of proteins
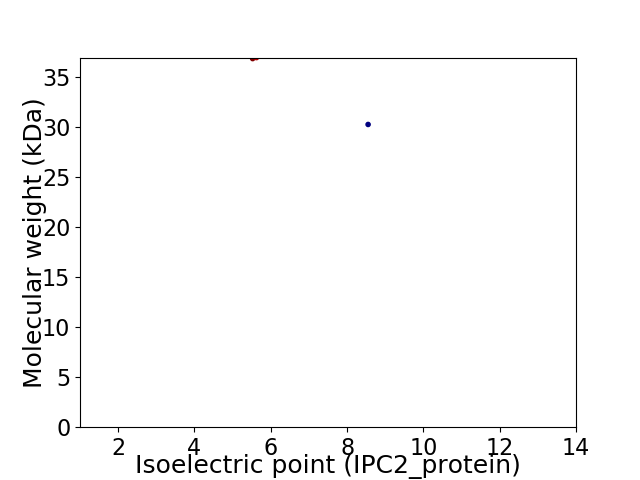
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L6KWB5|A0A1L6KWB5_9VIRU Capsid protein OS=Chicken associated smacovirus OX=1932006 GN=CAP PE=4 SV=1
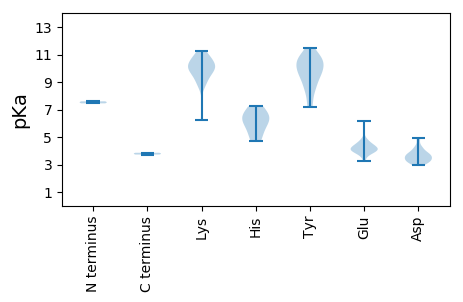
MM1 pKa = 7.6NSQTYY6 pKa = 10.45SYY8 pKa = 10.94FLDD11 pKa = 3.68LQTRR15 pKa = 11.84SDD17 pKa = 3.4RR18 pKa = 11.84VSVVKK23 pKa = 10.23VAAGGRR29 pKa = 11.84EE30 pKa = 3.84AMRR33 pKa = 11.84RR34 pKa = 11.84LLPFFAAYY42 pKa = 9.48KK43 pKa = 8.7YY44 pKa = 10.61FKK46 pKa = 10.36LGRR49 pKa = 11.84VKK51 pKa = 10.87LKK53 pKa = 9.78FVPASTLPVDD63 pKa = 3.64PTGLSLEE70 pKa = 4.56AGEE73 pKa = 4.25QTVDD77 pKa = 4.27PRR79 pKa = 11.84DD80 pKa = 3.35QFNPGLVRR88 pKa = 11.84ITNGEE93 pKa = 4.02YY94 pKa = 9.56WGNPQDD100 pKa = 4.81DD101 pKa = 4.19SQVTNMDD108 pKa = 3.26RR109 pKa = 11.84YY110 pKa = 8.59YY111 pKa = 11.3QLMLDD116 pKa = 3.72NRR118 pKa = 11.84WYY120 pKa = 10.83KK121 pKa = 10.86FNLQSGFEE129 pKa = 3.98RR130 pKa = 11.84HH131 pKa = 5.39ATPLYY136 pKa = 9.72WGIGQIRR143 pKa = 11.84QDD145 pKa = 3.81IFPGIAQNYY154 pKa = 7.3PGRR157 pKa = 11.84QAIGEE162 pKa = 4.4SEE164 pKa = 4.45VPQVISSKK172 pKa = 9.88TYY174 pKa = 9.45ISKK177 pKa = 10.37DD178 pKa = 3.15AQGSVKK184 pKa = 10.09TNLIWNSDD192 pKa = 3.28SSGWGIFQVGEE203 pKa = 4.06RR204 pKa = 11.84KK205 pKa = 9.67EE206 pKa = 4.85LGWLPTDD213 pKa = 3.45YY214 pKa = 10.76TLNEE218 pKa = 3.91QLGNFPGDD226 pKa = 3.2TGVNVPPEE234 pKa = 4.17IEE236 pKa = 4.55LLTIILPKK244 pKa = 10.11AYY246 pKa = 8.48KK247 pKa = 8.9TIYY250 pKa = 9.1YY251 pKa = 8.74YY252 pKa = 10.29RR253 pKa = 11.84VYY255 pKa = 9.82VTEE258 pKa = 4.23TVHH261 pKa = 7.23FKK263 pKa = 11.26DD264 pKa = 3.81PVAMNTVPMSDD275 pKa = 2.96STQLFNVGINGYY287 pKa = 10.24DD288 pKa = 3.43RR289 pKa = 11.84FWNVLAPGQASYY301 pKa = 11.44NVTPITSTQQYY312 pKa = 10.26NDD314 pKa = 3.19GDD316 pKa = 3.84GYY318 pKa = 10.93RR319 pKa = 11.84FRR321 pKa = 11.84EE322 pKa = 4.79GEE324 pKa = 3.86
MM1 pKa = 7.6NSQTYY6 pKa = 10.45SYY8 pKa = 10.94FLDD11 pKa = 3.68LQTRR15 pKa = 11.84SDD17 pKa = 3.4RR18 pKa = 11.84VSVVKK23 pKa = 10.23VAAGGRR29 pKa = 11.84EE30 pKa = 3.84AMRR33 pKa = 11.84RR34 pKa = 11.84LLPFFAAYY42 pKa = 9.48KK43 pKa = 8.7YY44 pKa = 10.61FKK46 pKa = 10.36LGRR49 pKa = 11.84VKK51 pKa = 10.87LKK53 pKa = 9.78FVPASTLPVDD63 pKa = 3.64PTGLSLEE70 pKa = 4.56AGEE73 pKa = 4.25QTVDD77 pKa = 4.27PRR79 pKa = 11.84DD80 pKa = 3.35QFNPGLVRR88 pKa = 11.84ITNGEE93 pKa = 4.02YY94 pKa = 9.56WGNPQDD100 pKa = 4.81DD101 pKa = 4.19SQVTNMDD108 pKa = 3.26RR109 pKa = 11.84YY110 pKa = 8.59YY111 pKa = 11.3QLMLDD116 pKa = 3.72NRR118 pKa = 11.84WYY120 pKa = 10.83KK121 pKa = 10.86FNLQSGFEE129 pKa = 3.98RR130 pKa = 11.84HH131 pKa = 5.39ATPLYY136 pKa = 9.72WGIGQIRR143 pKa = 11.84QDD145 pKa = 3.81IFPGIAQNYY154 pKa = 7.3PGRR157 pKa = 11.84QAIGEE162 pKa = 4.4SEE164 pKa = 4.45VPQVISSKK172 pKa = 9.88TYY174 pKa = 9.45ISKK177 pKa = 10.37DD178 pKa = 3.15AQGSVKK184 pKa = 10.09TNLIWNSDD192 pKa = 3.28SSGWGIFQVGEE203 pKa = 4.06RR204 pKa = 11.84KK205 pKa = 9.67EE206 pKa = 4.85LGWLPTDD213 pKa = 3.45YY214 pKa = 10.76TLNEE218 pKa = 3.91QLGNFPGDD226 pKa = 3.2TGVNVPPEE234 pKa = 4.17IEE236 pKa = 4.55LLTIILPKK244 pKa = 10.11AYY246 pKa = 8.48KK247 pKa = 8.9TIYY250 pKa = 9.1YY251 pKa = 8.74YY252 pKa = 10.29RR253 pKa = 11.84VYY255 pKa = 9.82VTEE258 pKa = 4.23TVHH261 pKa = 7.23FKK263 pKa = 11.26DD264 pKa = 3.81PVAMNTVPMSDD275 pKa = 2.96STQLFNVGINGYY287 pKa = 10.24DD288 pKa = 3.43RR289 pKa = 11.84FWNVLAPGQASYY301 pKa = 11.44NVTPITSTQQYY312 pKa = 10.26NDD314 pKa = 3.19GDD316 pKa = 3.84GYY318 pKa = 10.93RR319 pKa = 11.84FRR321 pKa = 11.84EE322 pKa = 4.79GEE324 pKa = 3.86
Molecular weight: 36.83 kDa
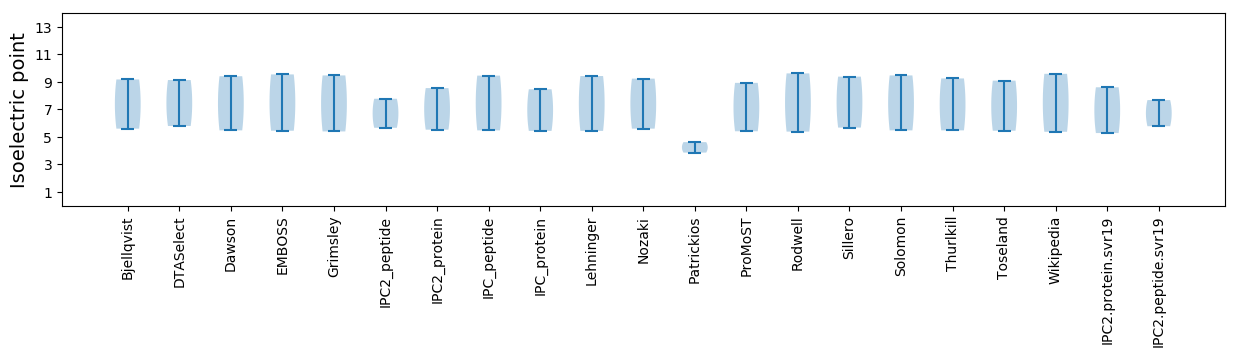
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L6KWB5|A0A1L6KWB5_9VIRU Capsid protein OS=Chicken associated smacovirus OX=1932006 GN=CAP PE=4 SV=1
MM1 pKa = 7.45NKK3 pKa = 9.26QAYY6 pKa = 8.55KK7 pKa = 10.82DD8 pKa = 3.66LGEE11 pKa = 4.36CQSMQTYY18 pKa = 9.37MLTIPRR24 pKa = 11.84KK25 pKa = 6.27VHH27 pKa = 5.94KK28 pKa = 9.22RR29 pKa = 11.84TLKK32 pKa = 10.49IMLEE36 pKa = 3.99QNDD39 pKa = 3.56VKK41 pKa = 10.91KK42 pKa = 11.11YY43 pKa = 10.31IIAKK47 pKa = 8.7EE48 pKa = 3.99RR49 pKa = 11.84GFGGYY54 pKa = 7.19EE55 pKa = 3.27HH56 pKa = 6.37WQIRR60 pKa = 11.84LKK62 pKa = 10.21TSNKK66 pKa = 9.82NFFIWCKK73 pKa = 10.02INIPEE78 pKa = 4.06AHH80 pKa = 6.39VEE82 pKa = 4.0EE83 pKa = 6.19AMDD86 pKa = 3.06TWDD89 pKa = 3.6YY90 pKa = 10.85EE91 pKa = 4.41RR92 pKa = 11.84KK93 pKa = 10.12EE94 pKa = 4.14GVYY97 pKa = 7.59WTSDD101 pKa = 3.2DD102 pKa = 3.82TNEE105 pKa = 3.59IRR107 pKa = 11.84ALRR110 pKa = 11.84FGKK113 pKa = 9.75PNLKK117 pKa = 9.56QKK119 pKa = 10.63RR120 pKa = 11.84VLKK123 pKa = 9.98LLKK126 pKa = 10.39YY127 pKa = 10.28QGDD130 pKa = 3.86RR131 pKa = 11.84NILVWYY137 pKa = 9.81DD138 pKa = 3.46PVGKK142 pKa = 10.12AGKK145 pKa = 9.25SWIVGHH151 pKa = 6.66LWEE154 pKa = 4.68QGKK157 pKa = 9.8ACYY160 pKa = 9.91VPPTLGTPKK169 pKa = 10.45EE170 pKa = 4.19IIQWVHH176 pKa = 4.72SAYY179 pKa = 10.67DD180 pKa = 3.63NEE182 pKa = 4.01GLIIIDD188 pKa = 4.92IPRR191 pKa = 11.84SWKK194 pKa = 9.57WNEE197 pKa = 3.71ALYY200 pKa = 9.3TAIEE204 pKa = 4.36TIKK207 pKa = 10.95DD208 pKa = 3.31GLVYY212 pKa = 10.41DD213 pKa = 4.16PRR215 pKa = 11.84YY216 pKa = 8.31SARR219 pKa = 11.84MKK221 pKa = 10.37NIRR224 pKa = 11.84GVKK227 pKa = 8.95VLVMTNTYY235 pKa = 9.8PRR237 pKa = 11.84VSALSEE243 pKa = 4.12DD244 pKa = 3.06RR245 pKa = 11.84WDD247 pKa = 4.21IINEE251 pKa = 4.0GGEE254 pKa = 4.21APLSS258 pKa = 3.72
MM1 pKa = 7.45NKK3 pKa = 9.26QAYY6 pKa = 8.55KK7 pKa = 10.82DD8 pKa = 3.66LGEE11 pKa = 4.36CQSMQTYY18 pKa = 9.37MLTIPRR24 pKa = 11.84KK25 pKa = 6.27VHH27 pKa = 5.94KK28 pKa = 9.22RR29 pKa = 11.84TLKK32 pKa = 10.49IMLEE36 pKa = 3.99QNDD39 pKa = 3.56VKK41 pKa = 10.91KK42 pKa = 11.11YY43 pKa = 10.31IIAKK47 pKa = 8.7EE48 pKa = 3.99RR49 pKa = 11.84GFGGYY54 pKa = 7.19EE55 pKa = 3.27HH56 pKa = 6.37WQIRR60 pKa = 11.84LKK62 pKa = 10.21TSNKK66 pKa = 9.82NFFIWCKK73 pKa = 10.02INIPEE78 pKa = 4.06AHH80 pKa = 6.39VEE82 pKa = 4.0EE83 pKa = 6.19AMDD86 pKa = 3.06TWDD89 pKa = 3.6YY90 pKa = 10.85EE91 pKa = 4.41RR92 pKa = 11.84KK93 pKa = 10.12EE94 pKa = 4.14GVYY97 pKa = 7.59WTSDD101 pKa = 3.2DD102 pKa = 3.82TNEE105 pKa = 3.59IRR107 pKa = 11.84ALRR110 pKa = 11.84FGKK113 pKa = 9.75PNLKK117 pKa = 9.56QKK119 pKa = 10.63RR120 pKa = 11.84VLKK123 pKa = 9.98LLKK126 pKa = 10.39YY127 pKa = 10.28QGDD130 pKa = 3.86RR131 pKa = 11.84NILVWYY137 pKa = 9.81DD138 pKa = 3.46PVGKK142 pKa = 10.12AGKK145 pKa = 9.25SWIVGHH151 pKa = 6.66LWEE154 pKa = 4.68QGKK157 pKa = 9.8ACYY160 pKa = 9.91VPPTLGTPKK169 pKa = 10.45EE170 pKa = 4.19IIQWVHH176 pKa = 4.72SAYY179 pKa = 10.67DD180 pKa = 3.63NEE182 pKa = 4.01GLIIIDD188 pKa = 4.92IPRR191 pKa = 11.84SWKK194 pKa = 9.57WNEE197 pKa = 3.71ALYY200 pKa = 9.3TAIEE204 pKa = 4.36TIKK207 pKa = 10.95DD208 pKa = 3.31GLVYY212 pKa = 10.41DD213 pKa = 4.16PRR215 pKa = 11.84YY216 pKa = 8.31SARR219 pKa = 11.84MKK221 pKa = 10.37NIRR224 pKa = 11.84GVKK227 pKa = 8.95VLVMTNTYY235 pKa = 9.8PRR237 pKa = 11.84VSALSEE243 pKa = 4.12DD244 pKa = 3.06RR245 pKa = 11.84WDD247 pKa = 4.21IINEE251 pKa = 4.0GGEE254 pKa = 4.21APLSS258 pKa = 3.72
Molecular weight: 30.25 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
582 |
258 |
324 |
291.0 |
33.54 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.811 ± 0.153 | 0.515 ± 0.434 |
5.67 ± 0.163 | 5.67 ± 0.876 |
3.265 ± 1.15 | 7.732 ± 0.766 |
1.203 ± 0.493 | 6.529 ± 1.34 |
6.529 ± 2.12 | 7.388 ± 0.016 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.234 ± 0.322 | 5.498 ± 0.308 |
5.326 ± 0.713 | 4.983 ± 1.002 |
5.67 ± 0.096 | 5.155 ± 0.858 |
6.014 ± 0.654 | 6.701 ± 0.595 |
3.093 ± 0.785 | 6.014 ± 0.394 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
