
Sphingobium cloacae
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Sphingomonadales; Sphingomonadaceae; Sphingobium
Average proteome isoelectric point is 6.72
Get precalculated fractions of proteins
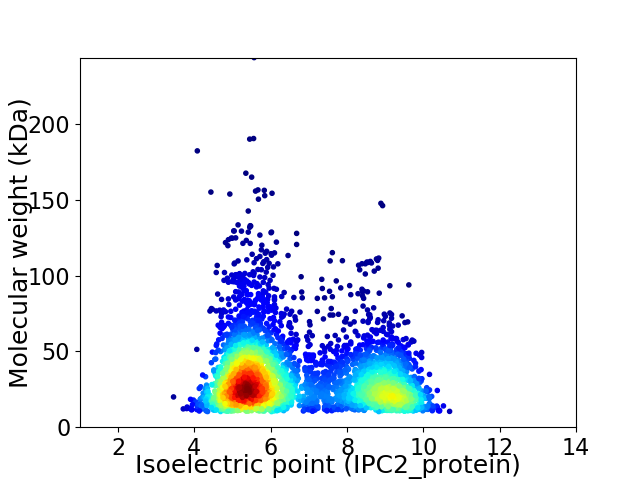
Virtual 2D-PAGE plot for 3852 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1E1F2D1|A0A1E1F2D1_9SPHN Flagellar protein FlgJ OS=Sphingobium cloacae OX=120107 GN=SCLO_1016370 PE=4 SV=1
MM1 pKa = 7.42TYY3 pKa = 10.78VVTDD7 pKa = 2.96NCIRR11 pKa = 11.84CKK13 pKa = 11.03YY14 pKa = 7.82MDD16 pKa = 4.38CVEE19 pKa = 4.32VCPVDD24 pKa = 3.79CFYY27 pKa = 11.19EE28 pKa = 4.58GEE30 pKa = 4.18NMLVINPSEE39 pKa = 4.67CIDD42 pKa = 4.07CGVCEE47 pKa = 4.44PEE49 pKa = 4.82CPAEE53 pKa = 5.13AILPDD58 pKa = 3.96TEE60 pKa = 4.78SGLEE64 pKa = 3.79SWLEE68 pKa = 3.84LNAKK72 pKa = 9.95YY73 pKa = 10.31SAEE76 pKa = 3.93WPNITVKK83 pKa = 10.86GDD85 pKa = 3.33APADD89 pKa = 3.78ADD91 pKa = 3.59EE92 pKa = 4.44MSGVEE97 pKa = 4.01NKK99 pKa = 10.26LEE101 pKa = 4.26KK102 pKa = 10.57FFSPNPGAGDD112 pKa = 3.39
MM1 pKa = 7.42TYY3 pKa = 10.78VVTDD7 pKa = 2.96NCIRR11 pKa = 11.84CKK13 pKa = 11.03YY14 pKa = 7.82MDD16 pKa = 4.38CVEE19 pKa = 4.32VCPVDD24 pKa = 3.79CFYY27 pKa = 11.19EE28 pKa = 4.58GEE30 pKa = 4.18NMLVINPSEE39 pKa = 4.67CIDD42 pKa = 4.07CGVCEE47 pKa = 4.44PEE49 pKa = 4.82CPAEE53 pKa = 5.13AILPDD58 pKa = 3.96TEE60 pKa = 4.78SGLEE64 pKa = 3.79SWLEE68 pKa = 3.84LNAKK72 pKa = 9.95YY73 pKa = 10.31SAEE76 pKa = 3.93WPNITVKK83 pKa = 10.86GDD85 pKa = 3.33APADD89 pKa = 3.78ADD91 pKa = 3.59EE92 pKa = 4.44MSGVEE97 pKa = 4.01NKK99 pKa = 10.26LEE101 pKa = 4.26KK102 pKa = 10.57FFSPNPGAGDD112 pKa = 3.39
Molecular weight: 12.28 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1E1EY52|A0A1E1EY52_9SPHN Acetate kinase OS=Sphingobium cloacae OX=120107 GN=ackA PE=3 SV=1
MM1 pKa = 6.92QVKK4 pKa = 9.42AHH6 pKa = 6.76KK7 pKa = 10.33LGLKK11 pKa = 7.83TAHH14 pKa = 7.05RR15 pKa = 11.84NPAPRR20 pKa = 11.84PRR22 pKa = 11.84LGGEE26 pKa = 4.13NLDD29 pKa = 3.38EE30 pKa = 5.17AIRR33 pKa = 11.84LRR35 pKa = 11.84EE36 pKa = 3.86VEE38 pKa = 4.0NWSFSAIGTHH48 pKa = 7.05FGICEE53 pKa = 3.73ASACNAVTIALCVRR67 pKa = 11.84RR68 pKa = 11.84GYY70 pKa = 10.62RR71 pKa = 11.84PAEE74 pKa = 4.07RR75 pKa = 11.84DD76 pKa = 3.0QHH78 pKa = 5.79GRR80 pKa = 11.84LTAEE84 pKa = 4.5GIEE87 pKa = 4.09RR88 pKa = 11.84LRR90 pKa = 11.84YY91 pKa = 9.26ALKK94 pKa = 10.45KK95 pKa = 9.73GYY97 pKa = 10.0KK98 pKa = 10.33GIDD101 pKa = 2.84IQLRR105 pKa = 11.84LGVSAACVSEE115 pKa = 3.49QRR117 pKa = 11.84RR118 pKa = 11.84RR119 pKa = 11.84YY120 pKa = 9.66NRR122 pKa = 11.84EE123 pKa = 3.08LLARR127 pKa = 11.84GKK129 pKa = 10.32AALPPPGGGEE139 pKa = 3.94AYY141 pKa = 10.52SGVKK145 pKa = 10.46LSPAKK150 pKa = 10.0RR151 pKa = 11.84RR152 pKa = 11.84QVEE155 pKa = 3.99EE156 pKa = 4.9LFLQGLGTQKK166 pKa = 9.78IAEE169 pKa = 4.16RR170 pKa = 11.84TGVSKK175 pKa = 9.39TSCTRR180 pKa = 11.84IRR182 pKa = 11.84GRR184 pKa = 11.84LIRR187 pKa = 11.84SLRR190 pKa = 11.84RR191 pKa = 11.84KK192 pKa = 10.22GEE194 pKa = 4.15SLPGCDD200 pKa = 2.98SCGVRR205 pKa = 11.84HH206 pKa = 5.43VHH208 pKa = 6.51AEE210 pKa = 3.79SARR213 pKa = 11.84FVTDD217 pKa = 3.21EE218 pKa = 4.18QKK220 pKa = 11.28DD221 pKa = 3.74LLRR224 pKa = 11.84AMLLDD229 pKa = 3.9RR230 pKa = 11.84VPVQRR235 pKa = 11.84AARR238 pKa = 11.84EE239 pKa = 3.99LAIGASTAYY248 pKa = 10.38RR249 pKa = 11.84LRR251 pKa = 11.84DD252 pKa = 3.39AFAAEE257 pKa = 3.96LAGEE261 pKa = 4.59GRR263 pKa = 11.84ALPPPRR269 pKa = 11.84RR270 pKa = 11.84PGRR273 pKa = 11.84VRR275 pKa = 11.84HH276 pKa = 5.42APMRR280 pKa = 11.84NSCWPPASPQEE291 pKa = 3.81IYY293 pKa = 10.99AFRR296 pKa = 11.84RR297 pKa = 11.84LLGCMGFAEE306 pKa = 5.8AKK308 pKa = 10.41AHH310 pKa = 5.14WQDD313 pKa = 2.95TRR315 pKa = 11.84RR316 pKa = 11.84EE317 pKa = 3.9EE318 pKa = 3.91ARR320 pKa = 11.84IARR323 pKa = 11.84EE324 pKa = 3.3AAATHH329 pKa = 6.64KK330 pKa = 10.21LTFEE334 pKa = 3.93EE335 pKa = 4.13QLAKK339 pKa = 10.42VASGEE344 pKa = 3.94LRR346 pKa = 11.84ITRR349 pKa = 11.84GFVRR353 pKa = 11.84NHH355 pKa = 6.55LEE357 pKa = 3.74PRR359 pKa = 11.84LPAQAVDD366 pKa = 3.37AA367 pKa = 4.95
MM1 pKa = 6.92QVKK4 pKa = 9.42AHH6 pKa = 6.76KK7 pKa = 10.33LGLKK11 pKa = 7.83TAHH14 pKa = 7.05RR15 pKa = 11.84NPAPRR20 pKa = 11.84PRR22 pKa = 11.84LGGEE26 pKa = 4.13NLDD29 pKa = 3.38EE30 pKa = 5.17AIRR33 pKa = 11.84LRR35 pKa = 11.84EE36 pKa = 3.86VEE38 pKa = 4.0NWSFSAIGTHH48 pKa = 7.05FGICEE53 pKa = 3.73ASACNAVTIALCVRR67 pKa = 11.84RR68 pKa = 11.84GYY70 pKa = 10.62RR71 pKa = 11.84PAEE74 pKa = 4.07RR75 pKa = 11.84DD76 pKa = 3.0QHH78 pKa = 5.79GRR80 pKa = 11.84LTAEE84 pKa = 4.5GIEE87 pKa = 4.09RR88 pKa = 11.84LRR90 pKa = 11.84YY91 pKa = 9.26ALKK94 pKa = 10.45KK95 pKa = 9.73GYY97 pKa = 10.0KK98 pKa = 10.33GIDD101 pKa = 2.84IQLRR105 pKa = 11.84LGVSAACVSEE115 pKa = 3.49QRR117 pKa = 11.84RR118 pKa = 11.84RR119 pKa = 11.84YY120 pKa = 9.66NRR122 pKa = 11.84EE123 pKa = 3.08LLARR127 pKa = 11.84GKK129 pKa = 10.32AALPPPGGGEE139 pKa = 3.94AYY141 pKa = 10.52SGVKK145 pKa = 10.46LSPAKK150 pKa = 10.0RR151 pKa = 11.84RR152 pKa = 11.84QVEE155 pKa = 3.99EE156 pKa = 4.9LFLQGLGTQKK166 pKa = 9.78IAEE169 pKa = 4.16RR170 pKa = 11.84TGVSKK175 pKa = 9.39TSCTRR180 pKa = 11.84IRR182 pKa = 11.84GRR184 pKa = 11.84LIRR187 pKa = 11.84SLRR190 pKa = 11.84RR191 pKa = 11.84KK192 pKa = 10.22GEE194 pKa = 4.15SLPGCDD200 pKa = 2.98SCGVRR205 pKa = 11.84HH206 pKa = 5.43VHH208 pKa = 6.51AEE210 pKa = 3.79SARR213 pKa = 11.84FVTDD217 pKa = 3.21EE218 pKa = 4.18QKK220 pKa = 11.28DD221 pKa = 3.74LLRR224 pKa = 11.84AMLLDD229 pKa = 3.9RR230 pKa = 11.84VPVQRR235 pKa = 11.84AARR238 pKa = 11.84EE239 pKa = 3.99LAIGASTAYY248 pKa = 10.38RR249 pKa = 11.84LRR251 pKa = 11.84DD252 pKa = 3.39AFAAEE257 pKa = 3.96LAGEE261 pKa = 4.59GRR263 pKa = 11.84ALPPPRR269 pKa = 11.84RR270 pKa = 11.84PGRR273 pKa = 11.84VRR275 pKa = 11.84HH276 pKa = 5.42APMRR280 pKa = 11.84NSCWPPASPQEE291 pKa = 3.81IYY293 pKa = 10.99AFRR296 pKa = 11.84RR297 pKa = 11.84LLGCMGFAEE306 pKa = 5.8AKK308 pKa = 10.41AHH310 pKa = 5.14WQDD313 pKa = 2.95TRR315 pKa = 11.84RR316 pKa = 11.84EE317 pKa = 3.9EE318 pKa = 3.91ARR320 pKa = 11.84IARR323 pKa = 11.84EE324 pKa = 3.3AAATHH329 pKa = 6.64KK330 pKa = 10.21LTFEE334 pKa = 3.93EE335 pKa = 4.13QLAKK339 pKa = 10.42VASGEE344 pKa = 3.94LRR346 pKa = 11.84ITRR349 pKa = 11.84GFVRR353 pKa = 11.84NHH355 pKa = 6.55LEE357 pKa = 3.74PRR359 pKa = 11.84LPAQAVDD366 pKa = 3.37AA367 pKa = 4.95
Molecular weight: 40.78 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1264410 |
99 |
2202 |
328.2 |
35.63 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.116 ± 0.057 | 0.781 ± 0.013 |
5.994 ± 0.031 | 5.571 ± 0.038 |
3.468 ± 0.024 | 8.696 ± 0.039 |
2.074 ± 0.02 | 5.087 ± 0.023 |
3.085 ± 0.032 | 10.031 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.559 ± 0.021 | 2.432 ± 0.025 |
5.283 ± 0.027 | 3.227 ± 0.023 |
7.802 ± 0.046 | 5.282 ± 0.029 |
4.98 ± 0.03 | 6.923 ± 0.032 |
1.427 ± 0.017 | 2.183 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
