
Blastococcus sp. TF02-9
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Geodermatophilales; Geodermatophilaceae; Blastococcus; unclassified Blastococcus
Average proteome isoelectric point is 6.04
Get precalculated fractions of proteins
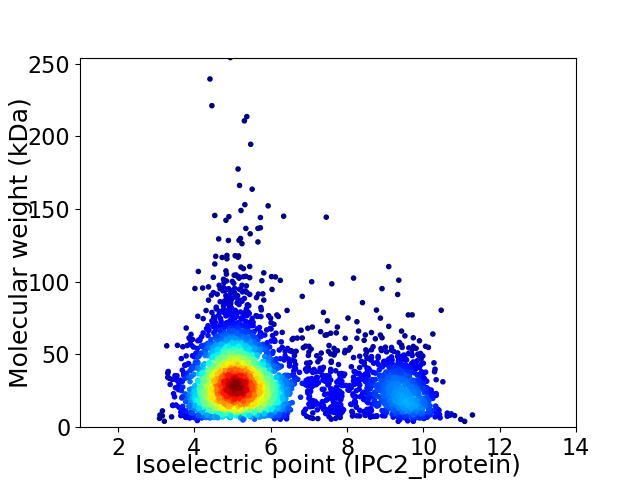
Virtual 2D-PAGE plot for 3851 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
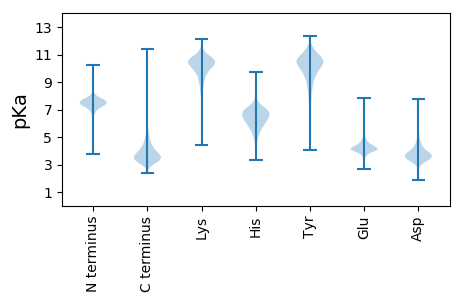
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A366ZEM5|A0A366ZEM5_9ACTN Isocitrate lyase/phosphoenolpyruvate mutase family protein OS=Blastococcus sp. TF02-9 OX=2250576 GN=DQ239_15900 PE=4 SV=1
MM1 pKa = 7.33NLGPVSRR8 pKa = 11.84SAAALAAVLLAATAGCSTKK27 pKa = 10.57APEE30 pKa = 4.14SSGGGGGGGDD40 pKa = 3.48AADD43 pKa = 3.63VQTDD47 pKa = 3.52VGVEE51 pKa = 3.98GTTIQLGVLTDD62 pKa = 3.65LTGVFAALGKK72 pKa = 10.36DD73 pKa = 3.2LTNANTLYY81 pKa = 10.3WEE83 pKa = 4.94DD84 pKa = 3.63NQVCDD89 pKa = 3.95TYY91 pKa = 11.53DD92 pKa = 3.34VEE94 pKa = 6.66LDD96 pKa = 3.83VQDD99 pKa = 3.57TGYY102 pKa = 10.9VPQTGVQLYY111 pKa = 10.71SGMKK115 pKa = 10.2DD116 pKa = 3.1SILAMQQTIGSPINTALAPEE136 pKa = 4.36YY137 pKa = 10.69EE138 pKa = 4.25SDD140 pKa = 3.81QIVNFPSAWSKK151 pKa = 9.42TLTEE155 pKa = 4.03IPGTGVVGATYY166 pKa = 10.42DD167 pKa = 3.59VEE169 pKa = 4.1ISNGYY174 pKa = 10.44DD175 pKa = 3.27YY176 pKa = 11.59LLQQGLLKK184 pKa = 10.96EE185 pKa = 4.43GDD187 pKa = 4.18TVGHH191 pKa = 6.94IYY193 pKa = 10.84FEE195 pKa = 4.56GEE197 pKa = 3.69YY198 pKa = 10.43GANGLAGSQAVAEE211 pKa = 4.29AKK213 pKa = 10.25GLNIVEE219 pKa = 4.25AQIKK223 pKa = 8.41STDD226 pKa = 3.31TDD228 pKa = 2.93MSAQVTQFKK237 pKa = 10.52AAGVKK242 pKa = 10.3AIALTVAPAQTTSVAAVAQAQGLDD266 pKa = 3.5VPMLGNNPVFAPGLLDD282 pKa = 4.95GPTADD287 pKa = 3.04WLKK290 pKa = 8.13THH292 pKa = 7.25LYY294 pKa = 8.55VASPVSSFADD304 pKa = 3.32HH305 pKa = 7.6ADD307 pKa = 3.68LLDD310 pKa = 4.93AYY312 pKa = 9.7QQAYY316 pKa = 9.51PDD318 pKa = 3.81VTPSLGVLVGYY329 pKa = 10.9GMATLMNQVLDD340 pKa = 4.18AACEE344 pKa = 4.07NGDD347 pKa = 3.68LTRR350 pKa = 11.84AGLVEE355 pKa = 4.64AFDD358 pKa = 4.06GLEE361 pKa = 4.19DD362 pKa = 3.77VDD364 pKa = 3.85TGGLVVPIRR373 pKa = 11.84GFEE376 pKa = 4.06NGKK379 pKa = 9.95SPSLEE384 pKa = 3.96SFVLVPADD392 pKa = 3.66EE393 pKa = 4.81PGGAKK398 pKa = 9.45VAQDD402 pKa = 3.22AFEE405 pKa = 5.06GEE407 pKa = 4.32FAADD411 pKa = 3.11IAGG414 pKa = 3.47
MM1 pKa = 7.33NLGPVSRR8 pKa = 11.84SAAALAAVLLAATAGCSTKK27 pKa = 10.57APEE30 pKa = 4.14SSGGGGGGGDD40 pKa = 3.48AADD43 pKa = 3.63VQTDD47 pKa = 3.52VGVEE51 pKa = 3.98GTTIQLGVLTDD62 pKa = 3.65LTGVFAALGKK72 pKa = 10.36DD73 pKa = 3.2LTNANTLYY81 pKa = 10.3WEE83 pKa = 4.94DD84 pKa = 3.63NQVCDD89 pKa = 3.95TYY91 pKa = 11.53DD92 pKa = 3.34VEE94 pKa = 6.66LDD96 pKa = 3.83VQDD99 pKa = 3.57TGYY102 pKa = 10.9VPQTGVQLYY111 pKa = 10.71SGMKK115 pKa = 10.2DD116 pKa = 3.1SILAMQQTIGSPINTALAPEE136 pKa = 4.36YY137 pKa = 10.69EE138 pKa = 4.25SDD140 pKa = 3.81QIVNFPSAWSKK151 pKa = 9.42TLTEE155 pKa = 4.03IPGTGVVGATYY166 pKa = 10.42DD167 pKa = 3.59VEE169 pKa = 4.1ISNGYY174 pKa = 10.44DD175 pKa = 3.27YY176 pKa = 11.59LLQQGLLKK184 pKa = 10.96EE185 pKa = 4.43GDD187 pKa = 4.18TVGHH191 pKa = 6.94IYY193 pKa = 10.84FEE195 pKa = 4.56GEE197 pKa = 3.69YY198 pKa = 10.43GANGLAGSQAVAEE211 pKa = 4.29AKK213 pKa = 10.25GLNIVEE219 pKa = 4.25AQIKK223 pKa = 8.41STDD226 pKa = 3.31TDD228 pKa = 2.93MSAQVTQFKK237 pKa = 10.52AAGVKK242 pKa = 10.3AIALTVAPAQTTSVAAVAQAQGLDD266 pKa = 3.5VPMLGNNPVFAPGLLDD282 pKa = 4.95GPTADD287 pKa = 3.04WLKK290 pKa = 8.13THH292 pKa = 7.25LYY294 pKa = 8.55VASPVSSFADD304 pKa = 3.32HH305 pKa = 7.6ADD307 pKa = 3.68LLDD310 pKa = 4.93AYY312 pKa = 9.7QQAYY316 pKa = 9.51PDD318 pKa = 3.81VTPSLGVLVGYY329 pKa = 10.9GMATLMNQVLDD340 pKa = 4.18AACEE344 pKa = 4.07NGDD347 pKa = 3.68LTRR350 pKa = 11.84AGLVEE355 pKa = 4.64AFDD358 pKa = 4.06GLEE361 pKa = 4.19DD362 pKa = 3.77VDD364 pKa = 3.85TGGLVVPIRR373 pKa = 11.84GFEE376 pKa = 4.06NGKK379 pKa = 9.95SPSLEE384 pKa = 3.96SFVLVPADD392 pKa = 3.66EE393 pKa = 4.81PGGAKK398 pKa = 9.45VAQDD402 pKa = 3.22AFEE405 pKa = 5.06GEE407 pKa = 4.32FAADD411 pKa = 3.11IAGG414 pKa = 3.47
Molecular weight: 42.49 kDa
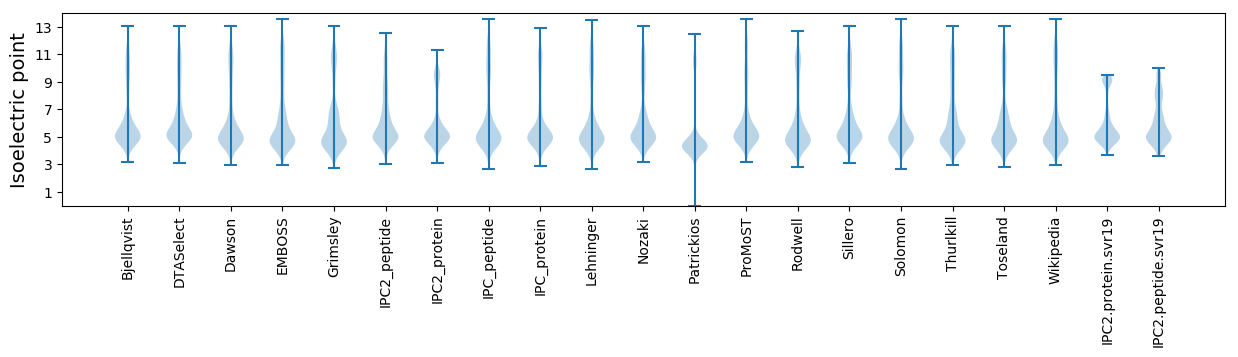
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A366ZVM9|A0A366ZVM9_9ACTN Type II toxin-antitoxin system PemK/MazF family toxin OS=Blastococcus sp. TF02-9 OX=2250576 GN=DQ239_00310 PE=3 SV=1
MM1 pKa = 7.68RR2 pKa = 11.84RR3 pKa = 11.84RR4 pKa = 11.84RR5 pKa = 11.84PGRR8 pKa = 11.84RR9 pKa = 11.84PRR11 pKa = 11.84ARR13 pKa = 11.84ARR15 pKa = 11.84PGGAGAGALPGRR27 pKa = 11.84RR28 pKa = 11.84ARR30 pKa = 11.84GGGAGRR36 pKa = 11.84RR37 pKa = 11.84GLAGLARR44 pKa = 11.84LALSSRR50 pKa = 11.84RR51 pKa = 11.84VMPHH55 pKa = 5.95PSLRR59 pKa = 11.84ARR61 pKa = 11.84PAQAHH66 pKa = 4.82RR67 pKa = 11.84RR68 pKa = 11.84IRR70 pKa = 11.84RR71 pKa = 11.84CRR73 pKa = 11.84PARR76 pKa = 11.84PP77 pKa = 3.31
MM1 pKa = 7.68RR2 pKa = 11.84RR3 pKa = 11.84RR4 pKa = 11.84RR5 pKa = 11.84PGRR8 pKa = 11.84RR9 pKa = 11.84PRR11 pKa = 11.84ARR13 pKa = 11.84ARR15 pKa = 11.84PGGAGAGALPGRR27 pKa = 11.84RR28 pKa = 11.84ARR30 pKa = 11.84GGGAGRR36 pKa = 11.84RR37 pKa = 11.84GLAGLARR44 pKa = 11.84LALSSRR50 pKa = 11.84RR51 pKa = 11.84VMPHH55 pKa = 5.95PSLRR59 pKa = 11.84ARR61 pKa = 11.84PAQAHH66 pKa = 4.82RR67 pKa = 11.84RR68 pKa = 11.84IRR70 pKa = 11.84RR71 pKa = 11.84CRR73 pKa = 11.84PARR76 pKa = 11.84PP77 pKa = 3.31
Molecular weight: 8.4 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1224882 |
32 |
2508 |
318.1 |
33.78 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.249 ± 0.061 | 0.679 ± 0.009 |
6.368 ± 0.032 | 5.69 ± 0.04 |
2.556 ± 0.024 | 9.574 ± 0.039 |
1.949 ± 0.018 | 2.88 ± 0.028 |
1.421 ± 0.025 | 10.639 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.622 ± 0.017 | 1.468 ± 0.016 |
6.182 ± 0.034 | 2.726 ± 0.022 |
8.167 ± 0.048 | 4.918 ± 0.027 |
5.988 ± 0.038 | 9.676 ± 0.04 |
1.484 ± 0.016 | 1.761 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
