
Acidihalobacter prosperus
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Chromatiales; Ectothiorhodospiraceae; Acidihalobacter
Average proteome isoelectric point is 6.81
Get precalculated fractions of proteins
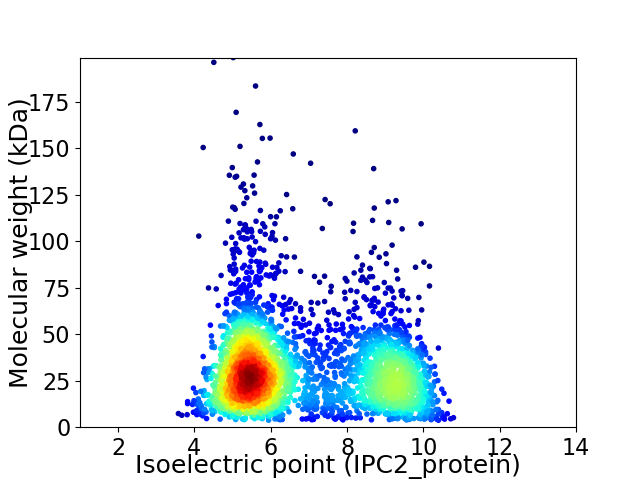
Virtual 2D-PAGE plot for 3188 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1A6C2A5|A0A1A6C2A5_9GAMM Uncharacterized protein OS=Acidihalobacter prosperus OX=160660 GN=Thpro_022946 PE=4 SV=1
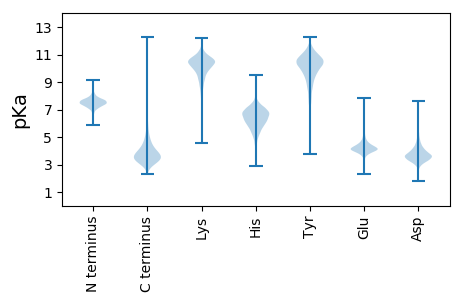
MM1 pKa = 6.38STEE4 pKa = 3.97VAEE7 pKa = 4.39NTMPDD12 pKa = 3.64PLLFTDD18 pKa = 4.27SAASKK23 pKa = 10.33VKK25 pKa = 10.41EE26 pKa = 5.1LIDD29 pKa = 4.16EE30 pKa = 4.46EE31 pKa = 4.58NNPNLKK37 pKa = 10.34LRR39 pKa = 11.84VFVTGGGCSGFQYY52 pKa = 11.01GFTFDD57 pKa = 4.44EE58 pKa = 4.41NVGDD62 pKa = 4.33GDD64 pKa = 4.11TTVEE68 pKa = 4.14NAGVTLLIDD77 pKa = 3.74PMSYY81 pKa = 10.31QYY83 pKa = 11.58LVGAEE88 pKa = 3.64IDD90 pKa = 3.75YY91 pKa = 10.81TEE93 pKa = 4.44GLEE96 pKa = 4.19GAQFVIRR103 pKa = 11.84NPNATTTCGCGSSFSVV119 pKa = 3.54
MM1 pKa = 6.38STEE4 pKa = 3.97VAEE7 pKa = 4.39NTMPDD12 pKa = 3.64PLLFTDD18 pKa = 4.27SAASKK23 pKa = 10.33VKK25 pKa = 10.41EE26 pKa = 5.1LIDD29 pKa = 4.16EE30 pKa = 4.46EE31 pKa = 4.58NNPNLKK37 pKa = 10.34LRR39 pKa = 11.84VFVTGGGCSGFQYY52 pKa = 11.01GFTFDD57 pKa = 4.44EE58 pKa = 4.41NVGDD62 pKa = 4.33GDD64 pKa = 4.11TTVEE68 pKa = 4.14NAGVTLLIDD77 pKa = 3.74PMSYY81 pKa = 10.31QYY83 pKa = 11.58LVGAEE88 pKa = 3.64IDD90 pKa = 3.75YY91 pKa = 10.81TEE93 pKa = 4.44GLEE96 pKa = 4.19GAQFVIRR103 pKa = 11.84NPNATTTCGCGSSFSVV119 pKa = 3.54
Molecular weight: 12.71 kDa
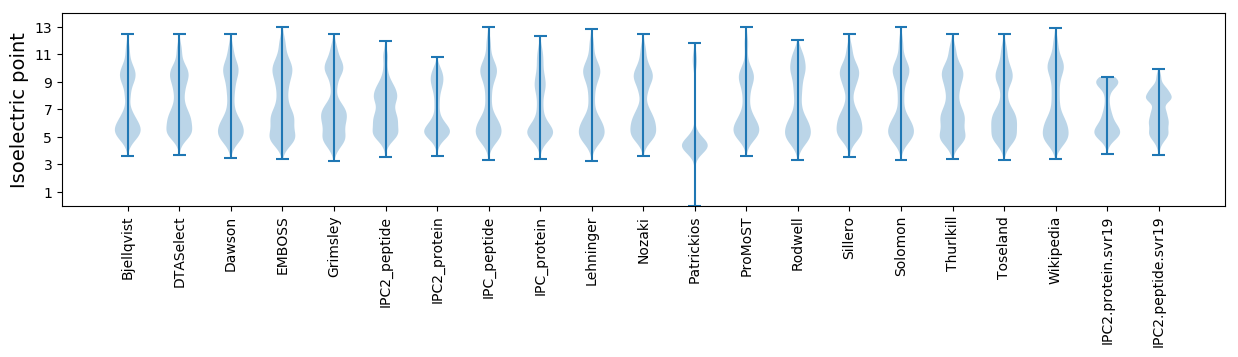
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1A6C4Q4|A0A1A6C4Q4_9GAMM Uncharacterized protein OS=Acidihalobacter prosperus OX=160660 GN=Thpro_021853 PE=3 SV=1
MM1 pKa = 7.14NAAGTSASVDD11 pKa = 3.53RR12 pKa = 11.84RR13 pKa = 11.84WPRR16 pKa = 11.84AIVLVDD22 pKa = 3.23MNAFFASIEE31 pKa = 3.99QLDD34 pKa = 4.04DD35 pKa = 3.71PSLRR39 pKa = 11.84GRR41 pKa = 11.84PVAVTNGDD49 pKa = 3.12QGTCIITCSYY59 pKa = 8.82EE60 pKa = 3.49ARR62 pKa = 11.84AFGIHH67 pKa = 5.62TGMRR71 pKa = 11.84LPEE74 pKa = 3.87ARR76 pKa = 11.84RR77 pKa = 11.84RR78 pKa = 11.84CPEE81 pKa = 4.82LIRR84 pKa = 11.84ITARR88 pKa = 11.84SARR91 pKa = 11.84YY92 pKa = 8.44AQISARR98 pKa = 11.84IMSALQAITPDD109 pKa = 3.1IEE111 pKa = 4.35VFSVDD116 pKa = 3.53EE117 pKa = 4.54AFLDD121 pKa = 3.66VTRR124 pKa = 11.84VQRR127 pKa = 11.84LHH129 pKa = 6.52GAPPVIAKK137 pKa = 9.62KK138 pKa = 7.86VQRR141 pKa = 11.84LVRR144 pKa = 11.84EE145 pKa = 4.28VSGLPCSVGVAGDD158 pKa = 3.3KK159 pKa = 7.75TTAKK163 pKa = 9.36WAAKK167 pKa = 7.74QRR169 pKa = 11.84KK170 pKa = 7.88PEE172 pKa = 3.98GVTVVPPWEE181 pKa = 3.81TAARR185 pKa = 11.84LRR187 pKa = 11.84NVPVTEE193 pKa = 3.93LCGVGRR199 pKa = 11.84GIGSFLASRR208 pKa = 11.84GVQTCGDD215 pKa = 3.9MARR218 pKa = 11.84LPVSVLAQRR227 pKa = 11.84WGNVGRR233 pKa = 11.84RR234 pKa = 11.84IWLMAQGRR242 pKa = 11.84DD243 pKa = 3.77PEE245 pKa = 4.71GVSPAVAPPKK255 pKa = 10.53SVGHH259 pKa = 5.76GKK261 pKa = 9.46ILPPATRR268 pKa = 11.84DD269 pKa = 3.4PRR271 pKa = 11.84LLQQYY276 pKa = 8.99LEE278 pKa = 4.46HH279 pKa = 6.59MAQKK283 pKa = 10.12VAARR287 pKa = 11.84LRR289 pKa = 11.84RR290 pKa = 11.84NGLEE294 pKa = 3.83AQQLFIGLRR303 pKa = 11.84TDD305 pKa = 3.46TGWLGGRR312 pKa = 11.84YY313 pKa = 7.53LTPRR317 pKa = 11.84PTDD320 pKa = 3.83DD321 pKa = 4.07GQPIAALCRR330 pKa = 11.84QVLARR335 pKa = 11.84VWQGEE340 pKa = 4.38GVHH343 pKa = 5.14QVQVTALDD351 pKa = 3.91PAPADD356 pKa = 3.51VQLDD360 pKa = 3.88LFEE363 pKa = 6.54DD364 pKa = 4.49PVAPTRR370 pKa = 11.84QAVNRR375 pKa = 11.84AVDD378 pKa = 4.56RR379 pKa = 11.84INRR382 pKa = 11.84RR383 pKa = 11.84FGGQAVATASLLGRR397 pKa = 11.84VPLHH401 pKa = 6.99DD402 pKa = 4.76VIAPAWKK409 pKa = 9.06PDD411 pKa = 3.79GIRR414 pKa = 11.84RR415 pKa = 11.84TVV417 pKa = 2.61
MM1 pKa = 7.14NAAGTSASVDD11 pKa = 3.53RR12 pKa = 11.84RR13 pKa = 11.84WPRR16 pKa = 11.84AIVLVDD22 pKa = 3.23MNAFFASIEE31 pKa = 3.99QLDD34 pKa = 4.04DD35 pKa = 3.71PSLRR39 pKa = 11.84GRR41 pKa = 11.84PVAVTNGDD49 pKa = 3.12QGTCIITCSYY59 pKa = 8.82EE60 pKa = 3.49ARR62 pKa = 11.84AFGIHH67 pKa = 5.62TGMRR71 pKa = 11.84LPEE74 pKa = 3.87ARR76 pKa = 11.84RR77 pKa = 11.84RR78 pKa = 11.84CPEE81 pKa = 4.82LIRR84 pKa = 11.84ITARR88 pKa = 11.84SARR91 pKa = 11.84YY92 pKa = 8.44AQISARR98 pKa = 11.84IMSALQAITPDD109 pKa = 3.1IEE111 pKa = 4.35VFSVDD116 pKa = 3.53EE117 pKa = 4.54AFLDD121 pKa = 3.66VTRR124 pKa = 11.84VQRR127 pKa = 11.84LHH129 pKa = 6.52GAPPVIAKK137 pKa = 9.62KK138 pKa = 7.86VQRR141 pKa = 11.84LVRR144 pKa = 11.84EE145 pKa = 4.28VSGLPCSVGVAGDD158 pKa = 3.3KK159 pKa = 7.75TTAKK163 pKa = 9.36WAAKK167 pKa = 7.74QRR169 pKa = 11.84KK170 pKa = 7.88PEE172 pKa = 3.98GVTVVPPWEE181 pKa = 3.81TAARR185 pKa = 11.84LRR187 pKa = 11.84NVPVTEE193 pKa = 3.93LCGVGRR199 pKa = 11.84GIGSFLASRR208 pKa = 11.84GVQTCGDD215 pKa = 3.9MARR218 pKa = 11.84LPVSVLAQRR227 pKa = 11.84WGNVGRR233 pKa = 11.84RR234 pKa = 11.84IWLMAQGRR242 pKa = 11.84DD243 pKa = 3.77PEE245 pKa = 4.71GVSPAVAPPKK255 pKa = 10.53SVGHH259 pKa = 5.76GKK261 pKa = 9.46ILPPATRR268 pKa = 11.84DD269 pKa = 3.4PRR271 pKa = 11.84LLQQYY276 pKa = 8.99LEE278 pKa = 4.46HH279 pKa = 6.59MAQKK283 pKa = 10.12VAARR287 pKa = 11.84LRR289 pKa = 11.84RR290 pKa = 11.84NGLEE294 pKa = 3.83AQQLFIGLRR303 pKa = 11.84TDD305 pKa = 3.46TGWLGGRR312 pKa = 11.84YY313 pKa = 7.53LTPRR317 pKa = 11.84PTDD320 pKa = 3.83DD321 pKa = 4.07GQPIAALCRR330 pKa = 11.84QVLARR335 pKa = 11.84VWQGEE340 pKa = 4.38GVHH343 pKa = 5.14QVQVTALDD351 pKa = 3.91PAPADD356 pKa = 3.51VQLDD360 pKa = 3.88LFEE363 pKa = 6.54DD364 pKa = 4.49PVAPTRR370 pKa = 11.84QAVNRR375 pKa = 11.84AVDD378 pKa = 4.56RR379 pKa = 11.84INRR382 pKa = 11.84RR383 pKa = 11.84FGGQAVATASLLGRR397 pKa = 11.84VPLHH401 pKa = 6.99DD402 pKa = 4.76VIAPAWKK409 pKa = 9.06PDD411 pKa = 3.79GIRR414 pKa = 11.84RR415 pKa = 11.84TVV417 pKa = 2.61
Molecular weight: 45.38 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
984843 |
37 |
1806 |
308.9 |
33.77 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.369 ± 0.062 | 0.884 ± 0.015 |
5.519 ± 0.035 | 5.652 ± 0.055 |
3.308 ± 0.028 | 8.504 ± 0.041 |
2.59 ± 0.023 | 4.671 ± 0.032 |
2.545 ± 0.034 | 11.287 ± 0.057 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.381 ± 0.022 | 2.45 ± 0.028 |
5.224 ± 0.034 | 3.561 ± 0.03 |
7.972 ± 0.044 | 5.009 ± 0.03 |
4.838 ± 0.031 | 7.212 ± 0.034 |
1.402 ± 0.02 | 2.623 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
