
Bovine papillomavirus type 5
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Epsilonpapillomavirus
Average proteome isoelectric point is 7.14
Get precalculated fractions of proteins
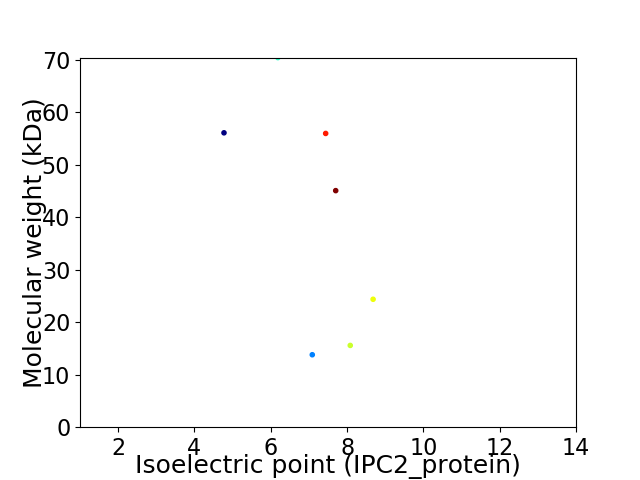
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q8BDG4|VE2_BPV5 Regulatory protein E2 OS=Bovine papillomavirus type 5 OX=40537 GN=E2 PE=3 SV=1
MM1 pKa = 7.53AVRR4 pKa = 11.84LRR6 pKa = 11.84RR7 pKa = 11.84VKK9 pKa = 10.35RR10 pKa = 11.84ANPYY14 pKa = 10.57DD15 pKa = 4.17LYY17 pKa = 9.98RR18 pKa = 11.84TCATGDD24 pKa = 3.64CPQDD28 pKa = 3.23VKK30 pKa = 11.53DD31 pKa = 3.89RR32 pKa = 11.84FEE34 pKa = 5.45HH35 pKa = 5.4NTIADD40 pKa = 4.66KK41 pKa = 10.59ILKK44 pKa = 8.59WGSAGVFLGGLGIGSTQARR63 pKa = 11.84PGLGTYY69 pKa = 10.29SPLGRR74 pKa = 11.84GGVTGRR80 pKa = 11.84IPVRR84 pKa = 11.84GPGSTRR90 pKa = 11.84PLGRR94 pKa = 11.84PFSSGPIDD102 pKa = 3.6TIGAGVRR109 pKa = 11.84TSVEE113 pKa = 4.01TSVTVPDD120 pKa = 4.04VVAVLPEE127 pKa = 4.29SPAVITPDD135 pKa = 3.88SMPVDD140 pKa = 4.08PGVGGLDD147 pKa = 3.01ISAEE151 pKa = 4.08IIEE154 pKa = 4.65EE155 pKa = 4.01PSLTFVEE162 pKa = 4.28PHH164 pKa = 6.23GPEE167 pKa = 4.02DD168 pKa = 4.12VAVLDD173 pKa = 4.23VNPAEE178 pKa = 4.84HH179 pKa = 7.09DD180 pKa = 3.46RR181 pKa = 11.84SVYY184 pKa = 10.38LSSSTTHH191 pKa = 6.51HH192 pKa = 6.23NPSFQGQVTVYY203 pKa = 9.67TDD205 pKa = 2.84IGEE208 pKa = 4.33TSEE211 pKa = 4.42TEE213 pKa = 3.7NLLISGSNIGSSRR226 pKa = 11.84GEE228 pKa = 4.17EE229 pKa = 3.54IQMQLFSGPKK239 pKa = 8.3TSTPEE244 pKa = 3.36TDD246 pKa = 3.01AVTKK250 pKa = 10.53VRR252 pKa = 11.84GRR254 pKa = 11.84ANWFSKK260 pKa = 10.24RR261 pKa = 11.84YY262 pKa = 6.12YY263 pKa = 8.41TQTSVRR269 pKa = 11.84DD270 pKa = 3.44PTFIQEE276 pKa = 4.09PQTYY280 pKa = 9.32FYY282 pKa = 11.25GFEE285 pKa = 4.1NPAYY289 pKa = 9.79EE290 pKa = 4.59PDD292 pKa = 3.74PFEE295 pKa = 6.54DD296 pKa = 3.81SFDD299 pKa = 3.72VQLASPSEE307 pKa = 4.02PVQPEE312 pKa = 3.94LRR314 pKa = 11.84DD315 pKa = 3.52ITHH318 pKa = 6.16VSAARR323 pKa = 11.84TFRR326 pKa = 11.84GEE328 pKa = 3.87SGRR331 pKa = 11.84VGISRR336 pKa = 11.84LGQKK340 pKa = 10.28SSIQTRR346 pKa = 11.84SGVTVGGRR354 pKa = 11.84VHH356 pKa = 6.78FRR358 pKa = 11.84YY359 pKa = 10.18SLSTIEE365 pKa = 5.63DD366 pKa = 4.37AIEE369 pKa = 4.1DD370 pKa = 3.6AGEE373 pKa = 4.2IEE375 pKa = 4.23LQVTNGSQGPSGSLQHH391 pKa = 5.73TAEE394 pKa = 4.74TILSEE399 pKa = 4.02GHH401 pKa = 6.66DD402 pKa = 3.81AYY404 pKa = 11.65VDD406 pKa = 3.29VDD408 pKa = 3.6MDD410 pKa = 4.51SVGSLYY416 pKa = 10.93SDD418 pKa = 3.2IDD420 pKa = 4.59LIDD423 pKa = 3.6EE424 pKa = 4.6HH425 pKa = 9.18SEE427 pKa = 4.23TPHH430 pKa = 6.79GILVFHH436 pKa = 7.53DD437 pKa = 3.81EE438 pKa = 4.54AEE440 pKa = 4.26TDD442 pKa = 3.88VVPVIDD448 pKa = 3.4VSYY451 pKa = 10.73VRR453 pKa = 11.84KK454 pKa = 8.86PLSTIPGSDD463 pKa = 3.37LWPTNINIQNGPVDD477 pKa = 4.07VDD479 pKa = 4.02LQDD482 pKa = 4.64SILPGIIITDD492 pKa = 3.59SGVDD496 pKa = 3.19GTYY499 pKa = 10.76FLNTYY504 pKa = 8.03LHH506 pKa = 6.92PSLHH510 pKa = 5.77KK511 pKa = 9.49RR512 pKa = 11.84KK513 pKa = 9.64KK514 pKa = 10.03RR515 pKa = 11.84RR516 pKa = 11.84FSS518 pKa = 3.23
MM1 pKa = 7.53AVRR4 pKa = 11.84LRR6 pKa = 11.84RR7 pKa = 11.84VKK9 pKa = 10.35RR10 pKa = 11.84ANPYY14 pKa = 10.57DD15 pKa = 4.17LYY17 pKa = 9.98RR18 pKa = 11.84TCATGDD24 pKa = 3.64CPQDD28 pKa = 3.23VKK30 pKa = 11.53DD31 pKa = 3.89RR32 pKa = 11.84FEE34 pKa = 5.45HH35 pKa = 5.4NTIADD40 pKa = 4.66KK41 pKa = 10.59ILKK44 pKa = 8.59WGSAGVFLGGLGIGSTQARR63 pKa = 11.84PGLGTYY69 pKa = 10.29SPLGRR74 pKa = 11.84GGVTGRR80 pKa = 11.84IPVRR84 pKa = 11.84GPGSTRR90 pKa = 11.84PLGRR94 pKa = 11.84PFSSGPIDD102 pKa = 3.6TIGAGVRR109 pKa = 11.84TSVEE113 pKa = 4.01TSVTVPDD120 pKa = 4.04VVAVLPEE127 pKa = 4.29SPAVITPDD135 pKa = 3.88SMPVDD140 pKa = 4.08PGVGGLDD147 pKa = 3.01ISAEE151 pKa = 4.08IIEE154 pKa = 4.65EE155 pKa = 4.01PSLTFVEE162 pKa = 4.28PHH164 pKa = 6.23GPEE167 pKa = 4.02DD168 pKa = 4.12VAVLDD173 pKa = 4.23VNPAEE178 pKa = 4.84HH179 pKa = 7.09DD180 pKa = 3.46RR181 pKa = 11.84SVYY184 pKa = 10.38LSSSTTHH191 pKa = 6.51HH192 pKa = 6.23NPSFQGQVTVYY203 pKa = 9.67TDD205 pKa = 2.84IGEE208 pKa = 4.33TSEE211 pKa = 4.42TEE213 pKa = 3.7NLLISGSNIGSSRR226 pKa = 11.84GEE228 pKa = 4.17EE229 pKa = 3.54IQMQLFSGPKK239 pKa = 8.3TSTPEE244 pKa = 3.36TDD246 pKa = 3.01AVTKK250 pKa = 10.53VRR252 pKa = 11.84GRR254 pKa = 11.84ANWFSKK260 pKa = 10.24RR261 pKa = 11.84YY262 pKa = 6.12YY263 pKa = 8.41TQTSVRR269 pKa = 11.84DD270 pKa = 3.44PTFIQEE276 pKa = 4.09PQTYY280 pKa = 9.32FYY282 pKa = 11.25GFEE285 pKa = 4.1NPAYY289 pKa = 9.79EE290 pKa = 4.59PDD292 pKa = 3.74PFEE295 pKa = 6.54DD296 pKa = 3.81SFDD299 pKa = 3.72VQLASPSEE307 pKa = 4.02PVQPEE312 pKa = 3.94LRR314 pKa = 11.84DD315 pKa = 3.52ITHH318 pKa = 6.16VSAARR323 pKa = 11.84TFRR326 pKa = 11.84GEE328 pKa = 3.87SGRR331 pKa = 11.84VGISRR336 pKa = 11.84LGQKK340 pKa = 10.28SSIQTRR346 pKa = 11.84SGVTVGGRR354 pKa = 11.84VHH356 pKa = 6.78FRR358 pKa = 11.84YY359 pKa = 10.18SLSTIEE365 pKa = 5.63DD366 pKa = 4.37AIEE369 pKa = 4.1DD370 pKa = 3.6AGEE373 pKa = 4.2IEE375 pKa = 4.23LQVTNGSQGPSGSLQHH391 pKa = 5.73TAEE394 pKa = 4.74TILSEE399 pKa = 4.02GHH401 pKa = 6.66DD402 pKa = 3.81AYY404 pKa = 11.65VDD406 pKa = 3.29VDD408 pKa = 3.6MDD410 pKa = 4.51SVGSLYY416 pKa = 10.93SDD418 pKa = 3.2IDD420 pKa = 4.59LIDD423 pKa = 3.6EE424 pKa = 4.6HH425 pKa = 9.18SEE427 pKa = 4.23TPHH430 pKa = 6.79GILVFHH436 pKa = 7.53DD437 pKa = 3.81EE438 pKa = 4.54AEE440 pKa = 4.26TDD442 pKa = 3.88VVPVIDD448 pKa = 3.4VSYY451 pKa = 10.73VRR453 pKa = 11.84KK454 pKa = 8.86PLSTIPGSDD463 pKa = 3.37LWPTNINIQNGPVDD477 pKa = 4.07VDD479 pKa = 4.02LQDD482 pKa = 4.64SILPGIIITDD492 pKa = 3.59SGVDD496 pKa = 3.19GTYY499 pKa = 10.76FLNTYY504 pKa = 8.03LHH506 pKa = 6.92PSLHH510 pKa = 5.77KK511 pKa = 9.49RR512 pKa = 11.84KK513 pKa = 9.64KK514 pKa = 10.03RR515 pKa = 11.84RR516 pKa = 11.84FSS518 pKa = 3.23
Molecular weight: 56.09 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|P50806|VL1_BPV5 Major capsid protein L1 OS=Bovine papillomavirus type 5 OX=40537 GN=L1 PE=3 SV=2
MM1 pKa = 7.42KK2 pKa = 10.41LVMLLRR8 pKa = 11.84PPPQTHH14 pKa = 6.86RR15 pKa = 11.84ADD17 pKa = 3.83TTDD20 pKa = 3.7GLPGLQEE27 pKa = 4.02EE28 pKa = 4.84PRR30 pKa = 11.84GGDD33 pKa = 3.38GPTCSGPAPAIPDD46 pKa = 3.63SPSRR50 pKa = 11.84CLSRR54 pKa = 11.84GFAGRR59 pKa = 11.84DD60 pKa = 3.49PGCHH64 pKa = 5.94RR65 pKa = 11.84NRR67 pKa = 11.84HH68 pKa = 5.06RR69 pKa = 11.84VHH71 pKa = 7.06PYY73 pKa = 9.33ILSGGQRR80 pKa = 11.84ILVTSSSSSTVQGPLSSGSSQHH102 pKa = 5.05SQSRR106 pKa = 11.84GRR108 pKa = 11.84PPSPDD113 pKa = 2.95STEE116 pKa = 4.0TEE118 pKa = 4.12RR119 pKa = 11.84ARR121 pKa = 11.84TPVNSDD127 pKa = 3.05RR128 pKa = 11.84QRR130 pKa = 11.84AGEE133 pKa = 3.86FDD135 pKa = 4.13LLKK138 pKa = 10.79GGCRR142 pKa = 11.84PCCLIEE148 pKa = 4.36GNGNKK153 pKa = 9.29VKK155 pKa = 10.69CLRR158 pKa = 11.84FRR160 pKa = 11.84LKK162 pKa = 10.15KK163 pKa = 8.19SHH165 pKa = 7.09RR166 pKa = 11.84SRR168 pKa = 11.84FLDD171 pKa = 2.91ITTTFWATGDD181 pKa = 3.83EE182 pKa = 4.54GSDD185 pKa = 3.23RR186 pKa = 11.84QGNGTILITFTDD198 pKa = 3.66TTQRR202 pKa = 11.84DD203 pKa = 3.79LFLGSVSIPGEE214 pKa = 3.67LSVRR218 pKa = 11.84RR219 pKa = 11.84ITISTDD225 pKa = 2.54
MM1 pKa = 7.42KK2 pKa = 10.41LVMLLRR8 pKa = 11.84PPPQTHH14 pKa = 6.86RR15 pKa = 11.84ADD17 pKa = 3.83TTDD20 pKa = 3.7GLPGLQEE27 pKa = 4.02EE28 pKa = 4.84PRR30 pKa = 11.84GGDD33 pKa = 3.38GPTCSGPAPAIPDD46 pKa = 3.63SPSRR50 pKa = 11.84CLSRR54 pKa = 11.84GFAGRR59 pKa = 11.84DD60 pKa = 3.49PGCHH64 pKa = 5.94RR65 pKa = 11.84NRR67 pKa = 11.84HH68 pKa = 5.06RR69 pKa = 11.84VHH71 pKa = 7.06PYY73 pKa = 9.33ILSGGQRR80 pKa = 11.84ILVTSSSSSTVQGPLSSGSSQHH102 pKa = 5.05SQSRR106 pKa = 11.84GRR108 pKa = 11.84PPSPDD113 pKa = 2.95STEE116 pKa = 4.0TEE118 pKa = 4.12RR119 pKa = 11.84ARR121 pKa = 11.84TPVNSDD127 pKa = 3.05RR128 pKa = 11.84QRR130 pKa = 11.84AGEE133 pKa = 3.86FDD135 pKa = 4.13LLKK138 pKa = 10.79GGCRR142 pKa = 11.84PCCLIEE148 pKa = 4.36GNGNKK153 pKa = 9.29VKK155 pKa = 10.69CLRR158 pKa = 11.84FRR160 pKa = 11.84LKK162 pKa = 10.15KK163 pKa = 8.19SHH165 pKa = 7.09RR166 pKa = 11.84SRR168 pKa = 11.84FLDD171 pKa = 2.91ITTTFWATGDD181 pKa = 3.83EE182 pKa = 4.54GSDD185 pKa = 3.23RR186 pKa = 11.84QGNGTILITFTDD198 pKa = 3.66TTQRR202 pKa = 11.84DD203 pKa = 3.79LFLGSVSIPGEE214 pKa = 3.67LSVRR218 pKa = 11.84RR219 pKa = 11.84ITISTDD225 pKa = 2.54
Molecular weight: 24.38 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2530 |
124 |
626 |
361.4 |
40.19 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.889 ± 0.678 | 2.609 ± 0.698 |
6.166 ± 0.303 | 5.178 ± 0.321 |
3.992 ± 0.4 | 7.51 ± 0.889 |
2.332 ± 0.325 | 4.111 ± 0.568 |
4.822 ± 0.877 | 8.735 ± 0.624 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.581 ± 0.338 | 3.913 ± 0.751 |
6.403 ± 0.79 | 4.743 ± 0.483 |
6.482 ± 0.836 | 8.656 ± 0.885 |
6.443 ± 0.796 | 6.126 ± 0.72 |
1.304 ± 0.277 | 3.004 ± 0.525 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
