
Klebsiella phage ST11-OXA245phi3.2
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Podoviridae; unclassified Podoviridae
Average proteome isoelectric point is 6.25
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 71 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
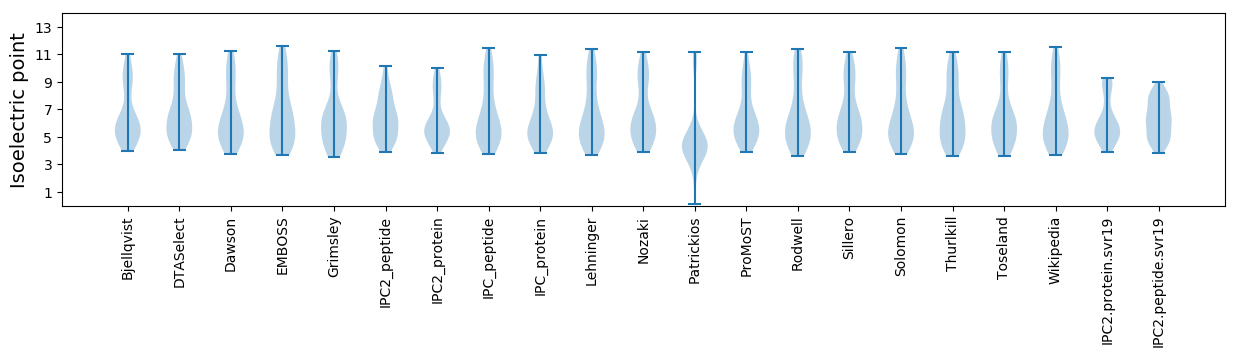
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A482MER0|A0A482MER0_9CAUD Uncharacterized protein OS=Klebsiella phage ST11-OXA245phi3.2 OX=2510477 PE=4 SV=1
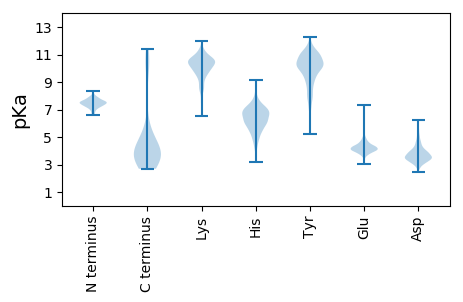
MM1 pKa = 7.68ANVDD5 pKa = 3.92PEE7 pKa = 4.56KK8 pKa = 9.89ITPVFALKK16 pKa = 10.0IVSIGEE22 pKa = 4.17VANGQCTLTLEE33 pKa = 4.49SGQDD37 pKa = 3.26VSDD40 pKa = 3.66PVVVTEE46 pKa = 5.48EE47 pKa = 4.13YY48 pKa = 9.78LQKK51 pKa = 10.65YY52 pKa = 9.34DD53 pKa = 4.05PQPGGYY59 pKa = 10.37YY60 pKa = 10.02IMCEE64 pKa = 3.94GGIGLYY70 pKa = 10.57SII72 pKa = 5.35
MM1 pKa = 7.68ANVDD5 pKa = 3.92PEE7 pKa = 4.56KK8 pKa = 9.89ITPVFALKK16 pKa = 10.0IVSIGEE22 pKa = 4.17VANGQCTLTLEE33 pKa = 4.49SGQDD37 pKa = 3.26VSDD40 pKa = 3.66PVVVTEE46 pKa = 5.48EE47 pKa = 4.13YY48 pKa = 9.78LQKK51 pKa = 10.65YY52 pKa = 9.34DD53 pKa = 4.05PQPGGYY59 pKa = 10.37YY60 pKa = 10.02IMCEE64 pKa = 3.94GGIGLYY70 pKa = 10.57SII72 pKa = 5.35
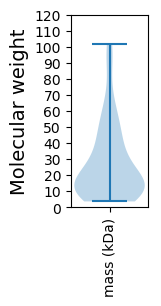
Molecular weight: 7.76 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A482MGQ9|A0A482MGQ9_9CAUD Nucleoside diphosphate kinase OS=Klebsiella phage ST11-OXA245phi3.2 OX=2510477 PE=3 SV=1
MM1 pKa = 7.51LPSQSPAIFTVSRR14 pKa = 11.84LNQTVRR20 pKa = 11.84LLLEE24 pKa = 4.18RR25 pKa = 11.84EE26 pKa = 4.17MGQVWISGEE35 pKa = 3.75ISNFSQPSSGHH46 pKa = 5.4WYY48 pKa = 7.89FTLKK52 pKa = 10.65DD53 pKa = 3.48DD54 pKa = 4.28NAQVRR59 pKa = 11.84CAMFRR64 pKa = 11.84NSNRR68 pKa = 11.84RR69 pKa = 11.84VTFRR73 pKa = 11.84PQHH76 pKa = 4.89GQQVLVRR83 pKa = 11.84ANITLYY89 pKa = 10.41EE90 pKa = 3.95PRR92 pKa = 11.84GDD94 pKa = 3.52YY95 pKa = 10.62QIIVEE100 pKa = 4.24SMQPAGEE107 pKa = 4.28GLLQQKK113 pKa = 9.56YY114 pKa = 7.98EE115 pKa = 3.96QLKK118 pKa = 10.07AQLTAEE124 pKa = 4.22GLFEE128 pKa = 4.22QKK130 pKa = 10.06HH131 pKa = 4.87KK132 pKa = 10.76QALPSPAHH140 pKa = 5.81CVGVITSKK148 pKa = 9.77TGAALHH154 pKa = 7.17DD155 pKa = 3.88ILHH158 pKa = 5.44VLRR161 pKa = 11.84RR162 pKa = 11.84RR163 pKa = 11.84DD164 pKa = 3.27PGLPVIIYY172 pKa = 6.6PTAVQGDD179 pKa = 4.28DD180 pKa = 3.52APGQIVRR187 pKa = 11.84AIALANARR195 pKa = 11.84QEE197 pKa = 4.1CDD199 pKa = 3.23VLIVGRR205 pKa = 11.84GGGSLEE211 pKa = 4.7DD212 pKa = 2.99LWSFNDD218 pKa = 3.26EE219 pKa = 3.92RR220 pKa = 11.84VARR223 pKa = 11.84AIFASQIPIVSAVGHH238 pKa = 5.46EE239 pKa = 4.25TDD241 pKa = 3.12VTIADD246 pKa = 4.4FVADD250 pKa = 4.65LRR252 pKa = 11.84APTPSAAAEE261 pKa = 3.88IVSRR265 pKa = 11.84NQQEE269 pKa = 4.85LLRR272 pKa = 11.84QLQSGQQRR280 pKa = 11.84LEE282 pKa = 3.56MAMDD286 pKa = 3.74YY287 pKa = 10.8FLASRR292 pKa = 11.84QRR294 pKa = 11.84RR295 pKa = 11.84FTQLFHH301 pKa = 7.77RR302 pKa = 11.84LQQQHH307 pKa = 5.81PQLRR311 pKa = 11.84LARR314 pKa = 11.84QQTALEE320 pKa = 4.14RR321 pKa = 11.84LRR323 pKa = 11.84QRR325 pKa = 11.84MRR327 pKa = 11.84IAVEE331 pKa = 4.13SQLKK335 pKa = 10.03RR336 pKa = 11.84AEE338 pKa = 3.99QRR340 pKa = 11.84QKK342 pKa = 10.35RR343 pKa = 11.84TVQRR347 pKa = 11.84LNHH350 pKa = 6.14YY351 pKa = 9.35NPQPRR356 pKa = 11.84IHH358 pKa = 7.04RR359 pKa = 11.84AQSRR363 pKa = 11.84IQQLEE368 pKa = 3.89YY369 pKa = 10.68RR370 pKa = 11.84LAEE373 pKa = 3.93IMRR376 pKa = 11.84GRR378 pKa = 11.84LSEE381 pKa = 3.84RR382 pKa = 11.84RR383 pKa = 11.84EE384 pKa = 3.89RR385 pKa = 11.84FGNAVTHH392 pKa = 6.54LEE394 pKa = 4.14AVSPLATLARR404 pKa = 11.84GYY406 pKa = 10.75SVTSVSDD413 pKa = 3.45GTVLKK418 pKa = 8.82QTKK421 pKa = 7.31QVKK424 pKa = 9.16TGDD427 pKa = 3.72LLTTRR432 pKa = 11.84LKK434 pKa = 11.04DD435 pKa = 3.13GWVEE439 pKa = 4.28SEE441 pKa = 4.33VKK443 pKa = 10.6QIATVKK449 pKa = 8.96KK450 pKa = 7.55TRR452 pKa = 11.84ARR454 pKa = 11.84KK455 pKa = 8.91PSPTKK460 pKa = 9.9PAEE463 pKa = 3.85
MM1 pKa = 7.51LPSQSPAIFTVSRR14 pKa = 11.84LNQTVRR20 pKa = 11.84LLLEE24 pKa = 4.18RR25 pKa = 11.84EE26 pKa = 4.17MGQVWISGEE35 pKa = 3.75ISNFSQPSSGHH46 pKa = 5.4WYY48 pKa = 7.89FTLKK52 pKa = 10.65DD53 pKa = 3.48DD54 pKa = 4.28NAQVRR59 pKa = 11.84CAMFRR64 pKa = 11.84NSNRR68 pKa = 11.84RR69 pKa = 11.84VTFRR73 pKa = 11.84PQHH76 pKa = 4.89GQQVLVRR83 pKa = 11.84ANITLYY89 pKa = 10.41EE90 pKa = 3.95PRR92 pKa = 11.84GDD94 pKa = 3.52YY95 pKa = 10.62QIIVEE100 pKa = 4.24SMQPAGEE107 pKa = 4.28GLLQQKK113 pKa = 9.56YY114 pKa = 7.98EE115 pKa = 3.96QLKK118 pKa = 10.07AQLTAEE124 pKa = 4.22GLFEE128 pKa = 4.22QKK130 pKa = 10.06HH131 pKa = 4.87KK132 pKa = 10.76QALPSPAHH140 pKa = 5.81CVGVITSKK148 pKa = 9.77TGAALHH154 pKa = 7.17DD155 pKa = 3.88ILHH158 pKa = 5.44VLRR161 pKa = 11.84RR162 pKa = 11.84RR163 pKa = 11.84DD164 pKa = 3.27PGLPVIIYY172 pKa = 6.6PTAVQGDD179 pKa = 4.28DD180 pKa = 3.52APGQIVRR187 pKa = 11.84AIALANARR195 pKa = 11.84QEE197 pKa = 4.1CDD199 pKa = 3.23VLIVGRR205 pKa = 11.84GGGSLEE211 pKa = 4.7DD212 pKa = 2.99LWSFNDD218 pKa = 3.26EE219 pKa = 3.92RR220 pKa = 11.84VARR223 pKa = 11.84AIFASQIPIVSAVGHH238 pKa = 5.46EE239 pKa = 4.25TDD241 pKa = 3.12VTIADD246 pKa = 4.4FVADD250 pKa = 4.65LRR252 pKa = 11.84APTPSAAAEE261 pKa = 3.88IVSRR265 pKa = 11.84NQQEE269 pKa = 4.85LLRR272 pKa = 11.84QLQSGQQRR280 pKa = 11.84LEE282 pKa = 3.56MAMDD286 pKa = 3.74YY287 pKa = 10.8FLASRR292 pKa = 11.84QRR294 pKa = 11.84RR295 pKa = 11.84FTQLFHH301 pKa = 7.77RR302 pKa = 11.84LQQQHH307 pKa = 5.81PQLRR311 pKa = 11.84LARR314 pKa = 11.84QQTALEE320 pKa = 4.14RR321 pKa = 11.84LRR323 pKa = 11.84QRR325 pKa = 11.84MRR327 pKa = 11.84IAVEE331 pKa = 4.13SQLKK335 pKa = 10.03RR336 pKa = 11.84AEE338 pKa = 3.99QRR340 pKa = 11.84QKK342 pKa = 10.35RR343 pKa = 11.84TVQRR347 pKa = 11.84LNHH350 pKa = 6.14YY351 pKa = 9.35NPQPRR356 pKa = 11.84IHH358 pKa = 7.04RR359 pKa = 11.84AQSRR363 pKa = 11.84IQQLEE368 pKa = 3.89YY369 pKa = 10.68RR370 pKa = 11.84LAEE373 pKa = 3.93IMRR376 pKa = 11.84GRR378 pKa = 11.84LSEE381 pKa = 3.84RR382 pKa = 11.84RR383 pKa = 11.84EE384 pKa = 3.89RR385 pKa = 11.84FGNAVTHH392 pKa = 6.54LEE394 pKa = 4.14AVSPLATLARR404 pKa = 11.84GYY406 pKa = 10.75SVTSVSDD413 pKa = 3.45GTVLKK418 pKa = 8.82QTKK421 pKa = 7.31QVKK424 pKa = 9.16TGDD427 pKa = 3.72LLTTRR432 pKa = 11.84LKK434 pKa = 11.04DD435 pKa = 3.13GWVEE439 pKa = 4.28SEE441 pKa = 4.33VKK443 pKa = 10.6QIATVKK449 pKa = 8.96KK450 pKa = 7.55TRR452 pKa = 11.84ARR454 pKa = 11.84KK455 pKa = 8.91PSPTKK460 pKa = 9.9PAEE463 pKa = 3.85
Molecular weight: 52.47 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
17912 |
31 |
943 |
252.3 |
27.84 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.362 ± 0.477 | 1.089 ± 0.127 |
6.158 ± 0.161 | 6.096 ± 0.287 |
3.199 ± 0.152 | 7.66 ± 0.247 |
1.764 ± 0.143 | 5.192 ± 0.187 |
4.578 ± 0.221 | 8.313 ± 0.257 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.697 ± 0.188 | 4.276 ± 0.237 |
4.5 ± 0.21 | 5.036 ± 0.338 |
6.23 ± 0.222 | 5.856 ± 0.233 |
5.84 ± 0.225 | 6.738 ± 0.332 |
1.502 ± 0.142 | 2.914 ± 0.156 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
