
Simian T-cell lymphotropic virus 4
Taxonomy: Viruses; Riboviria; Pararnavirae; Artverviricota; Revtraviricetes; Ortervirales; Retroviridae; Orthoretrovirinae; Lentivirus; Simian immunodeficiency virus
Average proteome isoelectric point is 8.05
Get precalculated fractions of proteins
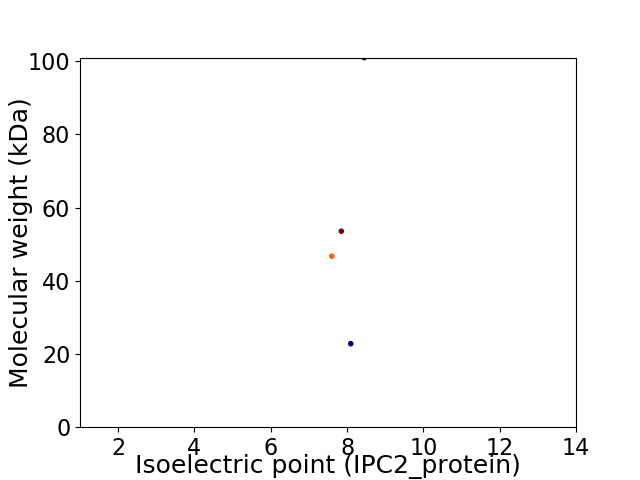
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|W5UB23|W5UB23_SIV Gag protein OS=Simian T-cell lymphotropic virus 4 OX=1454033 PE=4 SV=1
MM1 pKa = 7.18GQTHH5 pKa = 6.52TSSPVPKK12 pKa = 10.1APRR15 pKa = 11.84GLSTHH20 pKa = 5.79HH21 pKa = 6.88WLNFLQAAYY30 pKa = 9.95RR31 pKa = 11.84LQPGPSEE38 pKa = 4.33FDD40 pKa = 2.96FHH42 pKa = 6.44QLRR45 pKa = 11.84RR46 pKa = 11.84FLKK49 pKa = 10.6LALQTPVWLNPIDD62 pKa = 4.11YY63 pKa = 10.45SLLASLIPKK72 pKa = 8.97GYY74 pKa = 8.47PGRR77 pKa = 11.84VTEE80 pKa = 4.58IVNILLRR87 pKa = 11.84AHH89 pKa = 7.48PPPSAPAISMPSATGPAPAPQPQEE113 pKa = 3.66AHH115 pKa = 6.01TPPPYY120 pKa = 10.61AEE122 pKa = 4.25PAALQCLPIMHH133 pKa = 6.42SHH135 pKa = 6.84GAPSSHH141 pKa = 6.86RR142 pKa = 11.84PWQMKK147 pKa = 9.77DD148 pKa = 2.99LQAIKK153 pKa = 10.63QEE155 pKa = 4.28ISTSAPGSPQFMQTIRR171 pKa = 11.84LAIQQFDD178 pKa = 3.75PTAKK182 pKa = 10.22DD183 pKa = 3.33LHH185 pKa = 7.15DD186 pKa = 4.57LLQYY190 pKa = 10.65LCSSLIVSLHH200 pKa = 5.4HH201 pKa = 6.11QQLQALIAEE210 pKa = 4.59AEE212 pKa = 4.34TRR214 pKa = 11.84GLTGYY219 pKa = 10.95NPMAGPLRR227 pKa = 11.84VQANNPAQQGLRR239 pKa = 11.84RR240 pKa = 11.84EE241 pKa = 4.3YY242 pKa = 10.63QSLWLAAFAALPGNTRR258 pKa = 11.84DD259 pKa = 4.02PSWAAILQGLEE270 pKa = 3.8EE271 pKa = 4.83PYY273 pKa = 10.48CAFVEE278 pKa = 4.3RR279 pKa = 11.84LNVALDD285 pKa = 3.32NGLPEE290 pKa = 4.52GTPKK294 pKa = 10.76EE295 pKa = 4.23PILRR299 pKa = 11.84SLAYY303 pKa = 10.58SNANKK308 pKa = 9.97EE309 pKa = 4.36CQKK312 pKa = 10.72LLQARR317 pKa = 11.84GHH319 pKa = 5.59TNSPLGEE326 pKa = 4.11MLRR329 pKa = 11.84ACQAWTPKK337 pKa = 10.79DD338 pKa = 3.32KK339 pKa = 11.14TKK341 pKa = 11.11VLVVQPRR348 pKa = 11.84KK349 pKa = 8.11TPPTQPCFRR358 pKa = 11.84CGKK361 pKa = 8.35VGHH364 pKa = 6.73WSRR367 pKa = 11.84DD368 pKa = 3.52CTQPRR373 pKa = 11.84PPPGPCPLCQDD384 pKa = 3.51PSHH387 pKa = 7.13WKK389 pKa = 9.9RR390 pKa = 11.84DD391 pKa = 3.74CPQLKK396 pKa = 8.55TPPEE400 pKa = 4.06VEE402 pKa = 4.23EE403 pKa = 4.73PLLADD408 pKa = 5.0LPALLPEE415 pKa = 4.79EE416 pKa = 4.49KK417 pKa = 10.37NSPGGEE423 pKa = 3.95NN424 pKa = 3.13
MM1 pKa = 7.18GQTHH5 pKa = 6.52TSSPVPKK12 pKa = 10.1APRR15 pKa = 11.84GLSTHH20 pKa = 5.79HH21 pKa = 6.88WLNFLQAAYY30 pKa = 9.95RR31 pKa = 11.84LQPGPSEE38 pKa = 4.33FDD40 pKa = 2.96FHH42 pKa = 6.44QLRR45 pKa = 11.84RR46 pKa = 11.84FLKK49 pKa = 10.6LALQTPVWLNPIDD62 pKa = 4.11YY63 pKa = 10.45SLLASLIPKK72 pKa = 8.97GYY74 pKa = 8.47PGRR77 pKa = 11.84VTEE80 pKa = 4.58IVNILLRR87 pKa = 11.84AHH89 pKa = 7.48PPPSAPAISMPSATGPAPAPQPQEE113 pKa = 3.66AHH115 pKa = 6.01TPPPYY120 pKa = 10.61AEE122 pKa = 4.25PAALQCLPIMHH133 pKa = 6.42SHH135 pKa = 6.84GAPSSHH141 pKa = 6.86RR142 pKa = 11.84PWQMKK147 pKa = 9.77DD148 pKa = 2.99LQAIKK153 pKa = 10.63QEE155 pKa = 4.28ISTSAPGSPQFMQTIRR171 pKa = 11.84LAIQQFDD178 pKa = 3.75PTAKK182 pKa = 10.22DD183 pKa = 3.33LHH185 pKa = 7.15DD186 pKa = 4.57LLQYY190 pKa = 10.65LCSSLIVSLHH200 pKa = 5.4HH201 pKa = 6.11QQLQALIAEE210 pKa = 4.59AEE212 pKa = 4.34TRR214 pKa = 11.84GLTGYY219 pKa = 10.95NPMAGPLRR227 pKa = 11.84VQANNPAQQGLRR239 pKa = 11.84RR240 pKa = 11.84EE241 pKa = 4.3YY242 pKa = 10.63QSLWLAAFAALPGNTRR258 pKa = 11.84DD259 pKa = 4.02PSWAAILQGLEE270 pKa = 3.8EE271 pKa = 4.83PYY273 pKa = 10.48CAFVEE278 pKa = 4.3RR279 pKa = 11.84LNVALDD285 pKa = 3.32NGLPEE290 pKa = 4.52GTPKK294 pKa = 10.76EE295 pKa = 4.23PILRR299 pKa = 11.84SLAYY303 pKa = 10.58SNANKK308 pKa = 9.97EE309 pKa = 4.36CQKK312 pKa = 10.72LLQARR317 pKa = 11.84GHH319 pKa = 5.59TNSPLGEE326 pKa = 4.11MLRR329 pKa = 11.84ACQAWTPKK337 pKa = 10.79DD338 pKa = 3.32KK339 pKa = 11.14TKK341 pKa = 11.11VLVVQPRR348 pKa = 11.84KK349 pKa = 8.11TPPTQPCFRR358 pKa = 11.84CGKK361 pKa = 8.35VGHH364 pKa = 6.73WSRR367 pKa = 11.84DD368 pKa = 3.52CTQPRR373 pKa = 11.84PPPGPCPLCQDD384 pKa = 3.51PSHH387 pKa = 7.13WKK389 pKa = 9.9RR390 pKa = 11.84DD391 pKa = 3.74CPQLKK396 pKa = 8.55TPPEE400 pKa = 4.06VEE402 pKa = 4.23EE403 pKa = 4.73PLLADD408 pKa = 5.0LPALLPEE415 pKa = 4.79EE416 pKa = 4.49KK417 pKa = 10.37NSPGGEE423 pKa = 3.95NN424 pKa = 3.13
Molecular weight: 46.72 kDa
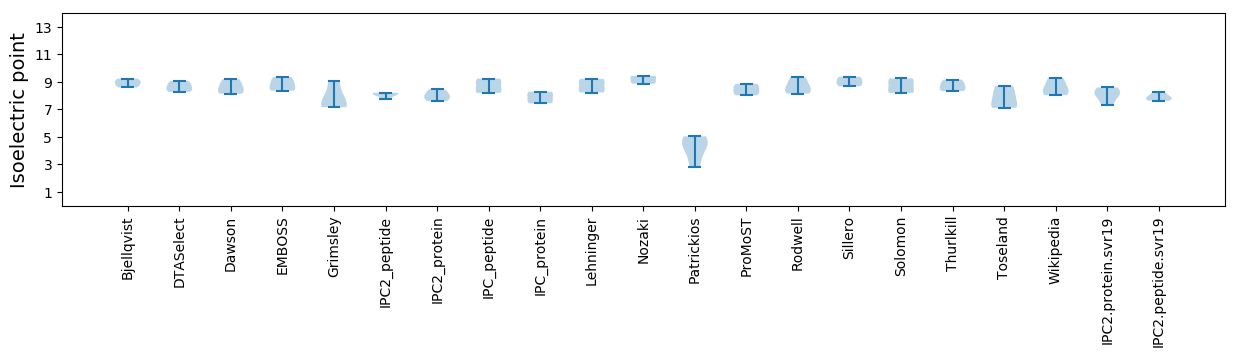
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|W5UB23|W5UB23_SIV Gag protein OS=Simian T-cell lymphotropic virus 4 OX=1454033 PE=4 SV=1
MM1 pKa = 7.9PGGPLLTGGPPGPDD15 pKa = 2.93PVIPGDD21 pKa = 3.96HH22 pKa = 6.83PCSHH26 pKa = 7.55RR27 pKa = 11.84LRR29 pKa = 11.84TSSRR33 pKa = 11.84APRR36 pKa = 11.84GQPVSFKK43 pKa = 10.47PEE45 pKa = 3.71RR46 pKa = 11.84LQALIDD52 pKa = 4.03LVSKK56 pKa = 10.73ALEE59 pKa = 4.32AGHH62 pKa = 6.72IEE64 pKa = 4.71PYY66 pKa = 10.27SGPGNNPVFPVKK78 pKa = 10.43KK79 pKa = 10.1PNGKK83 pKa = 8.28WRR85 pKa = 11.84FIHH88 pKa = 7.35DD89 pKa = 4.17LRR91 pKa = 11.84ATNAITTTLASPSPGPPDD109 pKa = 3.82LTSLPQALPHH119 pKa = 6.32LQTIDD124 pKa = 3.2LTDD127 pKa = 3.66AFFQIPLPKK136 pKa = 10.01RR137 pKa = 11.84FQPYY141 pKa = 9.14FAFTIPQPLNHH152 pKa = 6.83GPGSRR157 pKa = 11.84YY158 pKa = 9.81AWTVLPQGFKK168 pKa = 10.78NSPTLFEE175 pKa = 3.98QQLASVLGPARR186 pKa = 11.84KK187 pKa = 9.45AFPASVIVQYY197 pKa = 10.61MDD199 pKa = 6.41DD200 pKa = 4.5ILLACPSQHH209 pKa = 6.76EE210 pKa = 4.37LDD212 pKa = 3.73QLATLTAQLLSSHH225 pKa = 6.53GLPVSQEE232 pKa = 3.64KK233 pKa = 7.52TQRR236 pKa = 11.84TPGKK240 pKa = 9.59IHH242 pKa = 6.56FLGQIIHH249 pKa = 7.53PDD251 pKa = 3.42HH252 pKa = 6.45ITYY255 pKa = 7.62EE256 pKa = 4.28TTPTIPIKK264 pKa = 10.73AHH266 pKa = 4.78WTLTEE271 pKa = 3.96LQTLLGEE278 pKa = 4.4LQWVSKK284 pKa = 7.78GTPVLRR290 pKa = 11.84EE291 pKa = 4.21HH292 pKa = 6.65LHH294 pKa = 5.97CLYY297 pKa = 10.77SALRR301 pKa = 11.84GLKK304 pKa = 10.16DD305 pKa = 3.4PRR307 pKa = 11.84DD308 pKa = 4.36TITLRR313 pKa = 11.84HH314 pKa = 5.6PHH316 pKa = 6.1LHH318 pKa = 6.37ALHH321 pKa = 6.95NIQQALHH328 pKa = 5.96HH329 pKa = 6.16NCRR332 pKa = 11.84GRR334 pKa = 11.84LDD336 pKa = 3.41STLPLLGLIFLSPSGTTSVLFQTNHH361 pKa = 4.91KK362 pKa = 9.19WPLVWLHH369 pKa = 6.07TPHH372 pKa = 7.49PPTSLCPWGHH382 pKa = 6.46ILACTVLTLDD392 pKa = 4.64KK393 pKa = 11.05YY394 pKa = 11.06ALQHH398 pKa = 5.89YY399 pKa = 8.05GQLCKK404 pKa = 10.53SFHH407 pKa = 6.9HH408 pKa = 6.28NMSTQALHH416 pKa = 7.19DD417 pKa = 3.9FVKK420 pKa = 10.68NSSHH424 pKa = 6.87PSVAILIHH432 pKa = 6.0HH433 pKa = 6.34MHH435 pKa = 7.12RR436 pKa = 11.84FCDD439 pKa = 4.23LGRR442 pKa = 11.84QPPGPWRR449 pKa = 11.84TLLQLPALLRR459 pKa = 11.84EE460 pKa = 4.27PQLLRR465 pKa = 11.84PAFSLSPVVIDD476 pKa = 4.19QAPCLFSDD484 pKa = 5.36GSPQKK489 pKa = 10.3AAYY492 pKa = 9.03VIWDD496 pKa = 3.85KK497 pKa = 11.57VILSQRR503 pKa = 11.84SVPLPPHH510 pKa = 6.73ANNSAQKK517 pKa = 10.53GEE519 pKa = 4.23LVGLLLGLQAAQPWPSLNIFLDD541 pKa = 3.7SKK543 pKa = 10.79FLIRR547 pKa = 11.84YY548 pKa = 6.81LQSLASGAFQGSSTHH563 pKa = 6.27HH564 pKa = 6.97RR565 pKa = 11.84LQASLPTLLQGKK577 pKa = 8.07VVYY580 pKa = 10.03LHH582 pKa = 6.24HH583 pKa = 6.65TRR585 pKa = 11.84SHH587 pKa = 5.5TQLPDD592 pKa = 4.75PISTLNEE599 pKa = 4.05YY600 pKa = 9.83TDD602 pKa = 3.86SLIVAPVTPLKK613 pKa = 10.79PEE615 pKa = 4.08GLHH618 pKa = 7.2ALTHH622 pKa = 6.61CNQQALVSHH631 pKa = 6.93GATPAQAKK639 pKa = 9.57QLVQACRR646 pKa = 11.84TCQIINPQHH655 pKa = 5.99HH656 pKa = 6.55MPRR659 pKa = 11.84GHH661 pKa = 6.78IRR663 pKa = 11.84RR664 pKa = 11.84GHH666 pKa = 5.89FPNHH670 pKa = 4.51TWQGDD675 pKa = 3.81VTHH678 pKa = 7.23LKK680 pKa = 10.15HH681 pKa = 6.96KK682 pKa = 8.24RR683 pKa = 11.84TRR685 pKa = 11.84YY686 pKa = 9.31CLHH689 pKa = 5.81VWVDD693 pKa = 3.66TFSGAVSCVCKK704 pKa = 10.57KK705 pKa = 10.76KK706 pKa = 9.28EE707 pKa = 4.11TSSDD711 pKa = 3.8LIKK714 pKa = 10.12TLLHH718 pKa = 6.78AISVLGKK725 pKa = 8.83PFSVNTDD732 pKa = 2.95NGPAYY737 pKa = 10.6LSQEE741 pKa = 3.77FHH743 pKa = 6.62EE744 pKa = 5.68FCTTLCIKK752 pKa = 10.43HH753 pKa = 5.78STHH756 pKa = 6.28IPYY759 pKa = 10.59NPTSSGLVEE768 pKa = 4.14RR769 pKa = 11.84TNGILKK775 pKa = 8.95TLLYY779 pKa = 10.34KK780 pKa = 10.82YY781 pKa = 10.56FLDD784 pKa = 5.11HH785 pKa = 7.63PDD787 pKa = 4.09LPLEE791 pKa = 4.31SAVSKK796 pKa = 10.83ALWTINHH803 pKa = 6.43LNVMRR808 pKa = 11.84PCGKK812 pKa = 8.22TRR814 pKa = 11.84WQLHH818 pKa = 5.21HH819 pKa = 7.01TPPLPPISEE828 pKa = 4.79SIQTTPTGLHH838 pKa = 5.33WYY840 pKa = 9.23YY841 pKa = 11.24YY842 pKa = 7.96KK843 pKa = 10.59TPGLTNQRR851 pKa = 11.84WKK853 pKa = 11.34GPVQSLQEE861 pKa = 3.96AAGAALLQVSDD872 pKa = 5.82GSPQWIPWRR881 pKa = 11.84LLKK884 pKa = 9.93KK885 pKa = 7.83TVCPKK890 pKa = 10.44PDD892 pKa = 3.73DD893 pKa = 4.54PEE895 pKa = 4.41PAGHH899 pKa = 6.68VEE901 pKa = 4.13TDD903 pKa = 3.6HH904 pKa = 5.88QHH906 pKa = 6.15HH907 pKa = 6.59GG908 pKa = 3.53
MM1 pKa = 7.9PGGPLLTGGPPGPDD15 pKa = 2.93PVIPGDD21 pKa = 3.96HH22 pKa = 6.83PCSHH26 pKa = 7.55RR27 pKa = 11.84LRR29 pKa = 11.84TSSRR33 pKa = 11.84APRR36 pKa = 11.84GQPVSFKK43 pKa = 10.47PEE45 pKa = 3.71RR46 pKa = 11.84LQALIDD52 pKa = 4.03LVSKK56 pKa = 10.73ALEE59 pKa = 4.32AGHH62 pKa = 6.72IEE64 pKa = 4.71PYY66 pKa = 10.27SGPGNNPVFPVKK78 pKa = 10.43KK79 pKa = 10.1PNGKK83 pKa = 8.28WRR85 pKa = 11.84FIHH88 pKa = 7.35DD89 pKa = 4.17LRR91 pKa = 11.84ATNAITTTLASPSPGPPDD109 pKa = 3.82LTSLPQALPHH119 pKa = 6.32LQTIDD124 pKa = 3.2LTDD127 pKa = 3.66AFFQIPLPKK136 pKa = 10.01RR137 pKa = 11.84FQPYY141 pKa = 9.14FAFTIPQPLNHH152 pKa = 6.83GPGSRR157 pKa = 11.84YY158 pKa = 9.81AWTVLPQGFKK168 pKa = 10.78NSPTLFEE175 pKa = 3.98QQLASVLGPARR186 pKa = 11.84KK187 pKa = 9.45AFPASVIVQYY197 pKa = 10.61MDD199 pKa = 6.41DD200 pKa = 4.5ILLACPSQHH209 pKa = 6.76EE210 pKa = 4.37LDD212 pKa = 3.73QLATLTAQLLSSHH225 pKa = 6.53GLPVSQEE232 pKa = 3.64KK233 pKa = 7.52TQRR236 pKa = 11.84TPGKK240 pKa = 9.59IHH242 pKa = 6.56FLGQIIHH249 pKa = 7.53PDD251 pKa = 3.42HH252 pKa = 6.45ITYY255 pKa = 7.62EE256 pKa = 4.28TTPTIPIKK264 pKa = 10.73AHH266 pKa = 4.78WTLTEE271 pKa = 3.96LQTLLGEE278 pKa = 4.4LQWVSKK284 pKa = 7.78GTPVLRR290 pKa = 11.84EE291 pKa = 4.21HH292 pKa = 6.65LHH294 pKa = 5.97CLYY297 pKa = 10.77SALRR301 pKa = 11.84GLKK304 pKa = 10.16DD305 pKa = 3.4PRR307 pKa = 11.84DD308 pKa = 4.36TITLRR313 pKa = 11.84HH314 pKa = 5.6PHH316 pKa = 6.1LHH318 pKa = 6.37ALHH321 pKa = 6.95NIQQALHH328 pKa = 5.96HH329 pKa = 6.16NCRR332 pKa = 11.84GRR334 pKa = 11.84LDD336 pKa = 3.41STLPLLGLIFLSPSGTTSVLFQTNHH361 pKa = 4.91KK362 pKa = 9.19WPLVWLHH369 pKa = 6.07TPHH372 pKa = 7.49PPTSLCPWGHH382 pKa = 6.46ILACTVLTLDD392 pKa = 4.64KK393 pKa = 11.05YY394 pKa = 11.06ALQHH398 pKa = 5.89YY399 pKa = 8.05GQLCKK404 pKa = 10.53SFHH407 pKa = 6.9HH408 pKa = 6.28NMSTQALHH416 pKa = 7.19DD417 pKa = 3.9FVKK420 pKa = 10.68NSSHH424 pKa = 6.87PSVAILIHH432 pKa = 6.0HH433 pKa = 6.34MHH435 pKa = 7.12RR436 pKa = 11.84FCDD439 pKa = 4.23LGRR442 pKa = 11.84QPPGPWRR449 pKa = 11.84TLLQLPALLRR459 pKa = 11.84EE460 pKa = 4.27PQLLRR465 pKa = 11.84PAFSLSPVVIDD476 pKa = 4.19QAPCLFSDD484 pKa = 5.36GSPQKK489 pKa = 10.3AAYY492 pKa = 9.03VIWDD496 pKa = 3.85KK497 pKa = 11.57VILSQRR503 pKa = 11.84SVPLPPHH510 pKa = 6.73ANNSAQKK517 pKa = 10.53GEE519 pKa = 4.23LVGLLLGLQAAQPWPSLNIFLDD541 pKa = 3.7SKK543 pKa = 10.79FLIRR547 pKa = 11.84YY548 pKa = 6.81LQSLASGAFQGSSTHH563 pKa = 6.27HH564 pKa = 6.97RR565 pKa = 11.84LQASLPTLLQGKK577 pKa = 8.07VVYY580 pKa = 10.03LHH582 pKa = 6.24HH583 pKa = 6.65TRR585 pKa = 11.84SHH587 pKa = 5.5TQLPDD592 pKa = 4.75PISTLNEE599 pKa = 4.05YY600 pKa = 9.83TDD602 pKa = 3.86SLIVAPVTPLKK613 pKa = 10.79PEE615 pKa = 4.08GLHH618 pKa = 7.2ALTHH622 pKa = 6.61CNQQALVSHH631 pKa = 6.93GATPAQAKK639 pKa = 9.57QLVQACRR646 pKa = 11.84TCQIINPQHH655 pKa = 5.99HH656 pKa = 6.55MPRR659 pKa = 11.84GHH661 pKa = 6.78IRR663 pKa = 11.84RR664 pKa = 11.84GHH666 pKa = 5.89FPNHH670 pKa = 4.51TWQGDD675 pKa = 3.81VTHH678 pKa = 7.23LKK680 pKa = 10.15HH681 pKa = 6.96KK682 pKa = 8.24RR683 pKa = 11.84TRR685 pKa = 11.84YY686 pKa = 9.31CLHH689 pKa = 5.81VWVDD693 pKa = 3.66TFSGAVSCVCKK704 pKa = 10.57KK705 pKa = 10.76KK706 pKa = 9.28EE707 pKa = 4.11TSSDD711 pKa = 3.8LIKK714 pKa = 10.12TLLHH718 pKa = 6.78AISVLGKK725 pKa = 8.83PFSVNTDD732 pKa = 2.95NGPAYY737 pKa = 10.6LSQEE741 pKa = 3.77FHH743 pKa = 6.62EE744 pKa = 5.68FCTTLCIKK752 pKa = 10.43HH753 pKa = 5.78STHH756 pKa = 6.28IPYY759 pKa = 10.59NPTSSGLVEE768 pKa = 4.14RR769 pKa = 11.84TNGILKK775 pKa = 8.95TLLYY779 pKa = 10.34KK780 pKa = 10.82YY781 pKa = 10.56FLDD784 pKa = 5.11HH785 pKa = 7.63PDD787 pKa = 4.09LPLEE791 pKa = 4.31SAVSKK796 pKa = 10.83ALWTINHH803 pKa = 6.43LNVMRR808 pKa = 11.84PCGKK812 pKa = 8.22TRR814 pKa = 11.84WQLHH818 pKa = 5.21HH819 pKa = 7.01TPPLPPISEE828 pKa = 4.79SIQTTPTGLHH838 pKa = 5.33WYY840 pKa = 9.23YY841 pKa = 11.24YY842 pKa = 7.96KK843 pKa = 10.59TPGLTNQRR851 pKa = 11.84WKK853 pKa = 11.34GPVQSLQEE861 pKa = 3.96AAGAALLQVSDD872 pKa = 5.82GSPQWIPWRR881 pKa = 11.84LLKK884 pKa = 9.93KK885 pKa = 7.83TVCPKK890 pKa = 10.44PDD892 pKa = 3.73DD893 pKa = 4.54PEE895 pKa = 4.41PAGHH899 pKa = 6.68VEE901 pKa = 4.13TDD903 pKa = 3.6HH904 pKa = 5.88QHH906 pKa = 6.15HH907 pKa = 6.59GG908 pKa = 3.53
Molecular weight: 101.0 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2030 |
213 |
908 |
507.5 |
56.02 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.995 ± 0.837 | 2.463 ± 0.241 |
3.35 ± 0.177 | 3.054 ± 0.583 |
2.611 ± 0.262 | 5.764 ± 0.387 |
4.483 ± 1.133 | 4.286 ± 0.367 |
3.744 ± 0.416 | 13.547 ± 0.471 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
0.936 ± 0.248 | 3.005 ± 0.147 |
11.182 ± 1.005 | 6.355 ± 0.343 |
4.631 ± 0.31 | 7.734 ± 0.813 |
6.798 ± 0.647 | 4.631 ± 0.448 |
2.02 ± 0.207 | 2.414 ± 0.354 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
