
Rhizophagus sp. RF1 medium virus
Taxonomy: Viruses; Riboviria; dsRNA viruses; unclassified dsRNA viruses
Average proteome isoelectric point is 7.69
Get precalculated fractions of proteins
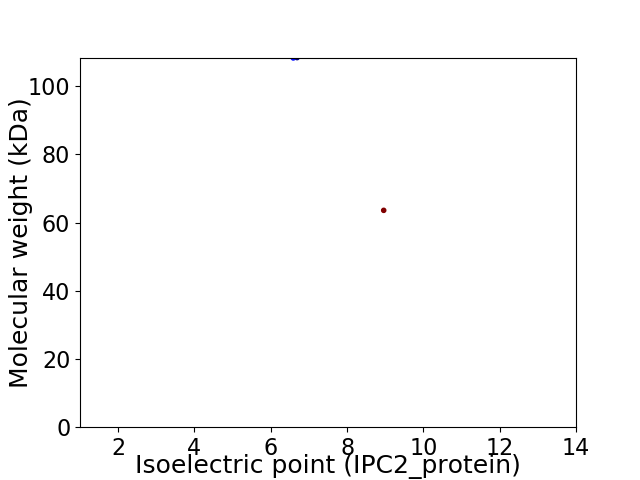
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|E2RWR2|E2RWR2_9VIRU Putative coat protein OS=Rhizophagus sp. RF1 medium virus OX=758865 GN=CP PE=4 SV=1
MM1 pKa = 8.01RR2 pKa = 11.84LPPFKK7 pKa = 9.8PDD9 pKa = 2.95GVIYY13 pKa = 10.3EE14 pKa = 4.55SYY16 pKa = 11.04CSLFNLNDD24 pKa = 3.24IKK26 pKa = 11.38DD27 pKa = 4.01VFNAPANFQIMKK39 pKa = 9.72YY40 pKa = 10.02EE41 pKa = 4.58PIFVNSRR48 pKa = 11.84PRR50 pKa = 11.84YY51 pKa = 9.26LYY53 pKa = 10.01PHH55 pKa = 7.24DD56 pKa = 3.63WDD58 pKa = 3.98YY59 pKa = 11.94TDD61 pKa = 4.46GAGAGQSIEE70 pKa = 4.26VNGVRR75 pKa = 11.84CFFSDD80 pKa = 3.88IYY82 pKa = 9.8SCRR85 pKa = 11.84WNRR88 pKa = 11.84FNLNLYY94 pKa = 9.56SDD96 pKa = 4.16SVKK99 pKa = 10.64LVNPKK104 pKa = 10.11SQTVDD109 pKa = 3.1MTTLTPEE116 pKa = 4.29DD117 pKa = 4.84LMDD120 pKa = 4.47IDD122 pKa = 4.18QDD124 pKa = 3.38NTRR127 pKa = 11.84IYY129 pKa = 11.14VFGRR133 pKa = 11.84LDD135 pKa = 3.24LSHH138 pKa = 7.75PYY140 pKa = 9.98FSLDD144 pKa = 3.52FPSLEE149 pKa = 4.11SAATKK154 pKa = 9.98IRR156 pKa = 11.84PSLYY160 pKa = 8.85FARR163 pKa = 11.84DD164 pKa = 3.16GRR166 pKa = 11.84RR167 pKa = 11.84LVVPILASAIGKK179 pKa = 9.88NYY181 pKa = 8.64LTYY184 pKa = 10.62QNIRR188 pKa = 11.84NVIYY192 pKa = 10.6GCLDD196 pKa = 3.37DD197 pKa = 5.96RR198 pKa = 11.84DD199 pKa = 4.35NLPEE203 pKa = 3.92VYY205 pKa = 9.8KK206 pKa = 11.06FNVSWVCQQPFLPQDD221 pKa = 3.11KK222 pKa = 10.94DD223 pKa = 3.67FFSFNQEE230 pKa = 3.26VGRR233 pKa = 11.84WCFVATVNEE242 pKa = 4.05NTPLHH247 pKa = 6.12IKK249 pKa = 10.57YY250 pKa = 8.17LTKK253 pKa = 10.86NLLDD257 pKa = 3.58QMRR260 pKa = 11.84KK261 pKa = 8.96QMTNDD266 pKa = 2.65SSLRR270 pKa = 11.84HH271 pKa = 4.85YY272 pKa = 10.94QSTEE276 pKa = 3.57LSHH279 pKa = 7.02LNHH282 pKa = 6.2EE283 pKa = 4.74QFANWVVKK291 pKa = 10.15VKK293 pKa = 10.62NIKK296 pKa = 9.76GLHH299 pKa = 5.61EE300 pKa = 4.33KK301 pKa = 8.81TWDD304 pKa = 3.33KK305 pKa = 11.36YY306 pKa = 11.18RR307 pKa = 11.84LGSDD311 pKa = 3.57SVIDD315 pKa = 3.61QYY317 pKa = 11.82VINYY321 pKa = 7.39DD322 pKa = 3.75TAIKK326 pKa = 9.99EE327 pKa = 4.12YY328 pKa = 10.52QKK330 pKa = 11.63LLGSYY335 pKa = 8.89VNKK338 pKa = 10.1EE339 pKa = 4.11RR340 pKa = 11.84IDD342 pKa = 3.47IDD344 pKa = 4.11SNLVDD349 pKa = 4.07EE350 pKa = 5.02LSGLGSVNEE359 pKa = 3.89KK360 pKa = 10.67VKK362 pKa = 10.26WDD364 pKa = 3.46RR365 pKa = 11.84CGDD368 pKa = 3.46NRR370 pKa = 11.84DD371 pKa = 3.64RR372 pKa = 11.84LPFKK376 pKa = 10.9DD377 pKa = 4.95DD378 pKa = 4.18FYY380 pKa = 11.88NPDD383 pKa = 3.22EE384 pKa = 4.46SGRR387 pKa = 11.84RR388 pKa = 11.84YY389 pKa = 10.41SHH391 pKa = 6.6VFMNPSFKK399 pKa = 10.56IFGVRR404 pKa = 11.84LQRR407 pKa = 11.84LLSMLDD413 pKa = 3.3CVIDD417 pKa = 4.06VKK419 pKa = 10.77HH420 pKa = 6.88DD421 pKa = 3.62KK422 pKa = 10.72YY423 pKa = 11.18LQDD426 pKa = 3.57VFSMVYY432 pKa = 9.9CVAEE436 pKa = 3.79VSRR439 pKa = 11.84RR440 pKa = 11.84FKK442 pKa = 11.25LLGDD446 pKa = 3.71FMKK449 pKa = 10.34KK450 pKa = 10.28YY451 pKa = 10.72GSNLTKK457 pKa = 10.46DD458 pKa = 2.79WYY460 pKa = 11.13VYY462 pKa = 11.25VNLEE466 pKa = 3.75TAIGYY471 pKa = 7.59KK472 pKa = 9.1TALPTDD478 pKa = 4.09EE479 pKa = 4.31FVHH482 pKa = 6.46MGGEE486 pKa = 4.2WIVGDD491 pKa = 4.23IEE493 pKa = 5.51HH494 pKa = 7.29IAPVPNGQLKK504 pKa = 9.45FYY506 pKa = 9.04EE507 pKa = 4.13WLVEE511 pKa = 4.55GIEE514 pKa = 4.09STLRR518 pKa = 11.84LKK520 pKa = 10.86GKK522 pKa = 10.14SKK524 pKa = 10.41IMYY527 pKa = 7.1EE528 pKa = 4.03KK529 pKa = 10.23PIEE532 pKa = 4.17FDD534 pKa = 3.03EE535 pKa = 5.18HH536 pKa = 6.13GNPMEE541 pKa = 4.3TLDD544 pKa = 3.54QFSYY548 pKa = 11.12DD549 pKa = 3.18VSKK552 pKa = 10.72QLSYY556 pKa = 11.0RR557 pKa = 11.84DD558 pKa = 3.67YY559 pKa = 11.45VYY561 pKa = 10.61GCTWSRR567 pKa = 11.84RR568 pKa = 11.84GSSSIKK574 pKa = 8.28TKK576 pKa = 10.65LRR578 pKa = 11.84AEE580 pKa = 4.11INGKK584 pKa = 8.31KK585 pKa = 10.22VKK587 pKa = 10.61LSEE590 pKa = 4.38TKK592 pKa = 9.17NTLAIYY598 pKa = 10.72GDD600 pKa = 3.85LEE602 pKa = 5.03KK603 pKa = 10.5ILKK606 pKa = 10.42DD607 pKa = 3.34SLDD610 pKa = 3.54PDD612 pKa = 3.71RR613 pKa = 11.84PQWGKK618 pKa = 10.81AIRR621 pKa = 11.84KK622 pKa = 9.16RR623 pKa = 11.84EE624 pKa = 3.87RR625 pKa = 11.84GKK627 pKa = 10.1IRR629 pKa = 11.84AVLGMDD635 pKa = 3.49VEE637 pKa = 5.31TYY639 pKa = 11.24LPMSYY644 pKa = 9.45IDD646 pKa = 6.28DD647 pKa = 4.91IIQPYY652 pKa = 10.11IIGNEE657 pKa = 3.96LSNLWEE663 pKa = 4.41TNFQRR668 pKa = 11.84HH669 pKa = 5.56AKK671 pKa = 10.3DD672 pKa = 2.96VDD674 pKa = 3.69RR675 pKa = 11.84LILSIEE681 pKa = 3.92KK682 pKa = 9.31TKK684 pKa = 10.52WFIPLDD690 pKa = 3.45QSGFDD695 pKa = 3.45HH696 pKa = 7.15QINNNMLLLTLVIIKK711 pKa = 9.84SWLLSRR717 pKa = 11.84VKK719 pKa = 10.96YY720 pKa = 10.51KK721 pKa = 11.11GDD723 pKa = 3.33VEE725 pKa = 4.25RR726 pKa = 11.84CIEE729 pKa = 4.2IIIKK733 pKa = 10.32RR734 pKa = 11.84LALTNSEE741 pKa = 4.41VNVDD745 pKa = 3.85DD746 pKa = 5.43KK747 pKa = 11.75INIPVEE753 pKa = 4.0RR754 pKa = 11.84GILSGWKK761 pKa = 7.04WTAFMDD767 pKa = 3.57TLINIGEE774 pKa = 4.36VNVALRR780 pKa = 11.84TCEE783 pKa = 3.91YY784 pKa = 10.62YY785 pKa = 11.23GLVCNYY791 pKa = 10.41KK792 pKa = 10.36LDD794 pKa = 4.0AQGDD798 pKa = 3.95DD799 pKa = 4.38DD800 pKa = 6.96DD801 pKa = 6.99IIIDD805 pKa = 4.15DD806 pKa = 4.76LLTGLVMCEE815 pKa = 3.49AYY817 pKa = 10.08RR818 pKa = 11.84YY819 pKa = 9.91CNLKK823 pKa = 10.23INPAKK828 pKa = 10.46FFIKK832 pKa = 10.74NNIDD836 pKa = 3.08EE837 pKa = 4.48YY838 pKa = 11.03LRR840 pKa = 11.84RR841 pKa = 11.84VYY843 pKa = 11.11VDD845 pKa = 4.0GVSLGYY851 pKa = 8.48PARR854 pKa = 11.84MFLTFFEE861 pKa = 5.1SNPIRR866 pKa = 11.84NVPLSEE872 pKa = 3.9MSEE875 pKa = 3.9LRR877 pKa = 11.84SVIKK881 pKa = 10.7NWMDD885 pKa = 4.77LIRR888 pKa = 11.84RR889 pKa = 11.84RR890 pKa = 11.84SLNNEE895 pKa = 3.47RR896 pKa = 11.84WVKK899 pKa = 10.02LVKK902 pKa = 10.67LMEE905 pKa = 4.75RR906 pKa = 11.84DD907 pKa = 2.89ILIRR911 pKa = 11.84VNKK914 pKa = 8.77NRR916 pKa = 11.84KK917 pKa = 8.77ISSLSLEE924 pKa = 4.57TILL927 pKa = 5.58
MM1 pKa = 8.01RR2 pKa = 11.84LPPFKK7 pKa = 9.8PDD9 pKa = 2.95GVIYY13 pKa = 10.3EE14 pKa = 4.55SYY16 pKa = 11.04CSLFNLNDD24 pKa = 3.24IKK26 pKa = 11.38DD27 pKa = 4.01VFNAPANFQIMKK39 pKa = 9.72YY40 pKa = 10.02EE41 pKa = 4.58PIFVNSRR48 pKa = 11.84PRR50 pKa = 11.84YY51 pKa = 9.26LYY53 pKa = 10.01PHH55 pKa = 7.24DD56 pKa = 3.63WDD58 pKa = 3.98YY59 pKa = 11.94TDD61 pKa = 4.46GAGAGQSIEE70 pKa = 4.26VNGVRR75 pKa = 11.84CFFSDD80 pKa = 3.88IYY82 pKa = 9.8SCRR85 pKa = 11.84WNRR88 pKa = 11.84FNLNLYY94 pKa = 9.56SDD96 pKa = 4.16SVKK99 pKa = 10.64LVNPKK104 pKa = 10.11SQTVDD109 pKa = 3.1MTTLTPEE116 pKa = 4.29DD117 pKa = 4.84LMDD120 pKa = 4.47IDD122 pKa = 4.18QDD124 pKa = 3.38NTRR127 pKa = 11.84IYY129 pKa = 11.14VFGRR133 pKa = 11.84LDD135 pKa = 3.24LSHH138 pKa = 7.75PYY140 pKa = 9.98FSLDD144 pKa = 3.52FPSLEE149 pKa = 4.11SAATKK154 pKa = 9.98IRR156 pKa = 11.84PSLYY160 pKa = 8.85FARR163 pKa = 11.84DD164 pKa = 3.16GRR166 pKa = 11.84RR167 pKa = 11.84LVVPILASAIGKK179 pKa = 9.88NYY181 pKa = 8.64LTYY184 pKa = 10.62QNIRR188 pKa = 11.84NVIYY192 pKa = 10.6GCLDD196 pKa = 3.37DD197 pKa = 5.96RR198 pKa = 11.84DD199 pKa = 4.35NLPEE203 pKa = 3.92VYY205 pKa = 9.8KK206 pKa = 11.06FNVSWVCQQPFLPQDD221 pKa = 3.11KK222 pKa = 10.94DD223 pKa = 3.67FFSFNQEE230 pKa = 3.26VGRR233 pKa = 11.84WCFVATVNEE242 pKa = 4.05NTPLHH247 pKa = 6.12IKK249 pKa = 10.57YY250 pKa = 8.17LTKK253 pKa = 10.86NLLDD257 pKa = 3.58QMRR260 pKa = 11.84KK261 pKa = 8.96QMTNDD266 pKa = 2.65SSLRR270 pKa = 11.84HH271 pKa = 4.85YY272 pKa = 10.94QSTEE276 pKa = 3.57LSHH279 pKa = 7.02LNHH282 pKa = 6.2EE283 pKa = 4.74QFANWVVKK291 pKa = 10.15VKK293 pKa = 10.62NIKK296 pKa = 9.76GLHH299 pKa = 5.61EE300 pKa = 4.33KK301 pKa = 8.81TWDD304 pKa = 3.33KK305 pKa = 11.36YY306 pKa = 11.18RR307 pKa = 11.84LGSDD311 pKa = 3.57SVIDD315 pKa = 3.61QYY317 pKa = 11.82VINYY321 pKa = 7.39DD322 pKa = 3.75TAIKK326 pKa = 9.99EE327 pKa = 4.12YY328 pKa = 10.52QKK330 pKa = 11.63LLGSYY335 pKa = 8.89VNKK338 pKa = 10.1EE339 pKa = 4.11RR340 pKa = 11.84IDD342 pKa = 3.47IDD344 pKa = 4.11SNLVDD349 pKa = 4.07EE350 pKa = 5.02LSGLGSVNEE359 pKa = 3.89KK360 pKa = 10.67VKK362 pKa = 10.26WDD364 pKa = 3.46RR365 pKa = 11.84CGDD368 pKa = 3.46NRR370 pKa = 11.84DD371 pKa = 3.64RR372 pKa = 11.84LPFKK376 pKa = 10.9DD377 pKa = 4.95DD378 pKa = 4.18FYY380 pKa = 11.88NPDD383 pKa = 3.22EE384 pKa = 4.46SGRR387 pKa = 11.84RR388 pKa = 11.84YY389 pKa = 10.41SHH391 pKa = 6.6VFMNPSFKK399 pKa = 10.56IFGVRR404 pKa = 11.84LQRR407 pKa = 11.84LLSMLDD413 pKa = 3.3CVIDD417 pKa = 4.06VKK419 pKa = 10.77HH420 pKa = 6.88DD421 pKa = 3.62KK422 pKa = 10.72YY423 pKa = 11.18LQDD426 pKa = 3.57VFSMVYY432 pKa = 9.9CVAEE436 pKa = 3.79VSRR439 pKa = 11.84RR440 pKa = 11.84FKK442 pKa = 11.25LLGDD446 pKa = 3.71FMKK449 pKa = 10.34KK450 pKa = 10.28YY451 pKa = 10.72GSNLTKK457 pKa = 10.46DD458 pKa = 2.79WYY460 pKa = 11.13VYY462 pKa = 11.25VNLEE466 pKa = 3.75TAIGYY471 pKa = 7.59KK472 pKa = 9.1TALPTDD478 pKa = 4.09EE479 pKa = 4.31FVHH482 pKa = 6.46MGGEE486 pKa = 4.2WIVGDD491 pKa = 4.23IEE493 pKa = 5.51HH494 pKa = 7.29IAPVPNGQLKK504 pKa = 9.45FYY506 pKa = 9.04EE507 pKa = 4.13WLVEE511 pKa = 4.55GIEE514 pKa = 4.09STLRR518 pKa = 11.84LKK520 pKa = 10.86GKK522 pKa = 10.14SKK524 pKa = 10.41IMYY527 pKa = 7.1EE528 pKa = 4.03KK529 pKa = 10.23PIEE532 pKa = 4.17FDD534 pKa = 3.03EE535 pKa = 5.18HH536 pKa = 6.13GNPMEE541 pKa = 4.3TLDD544 pKa = 3.54QFSYY548 pKa = 11.12DD549 pKa = 3.18VSKK552 pKa = 10.72QLSYY556 pKa = 11.0RR557 pKa = 11.84DD558 pKa = 3.67YY559 pKa = 11.45VYY561 pKa = 10.61GCTWSRR567 pKa = 11.84RR568 pKa = 11.84GSSSIKK574 pKa = 8.28TKK576 pKa = 10.65LRR578 pKa = 11.84AEE580 pKa = 4.11INGKK584 pKa = 8.31KK585 pKa = 10.22VKK587 pKa = 10.61LSEE590 pKa = 4.38TKK592 pKa = 9.17NTLAIYY598 pKa = 10.72GDD600 pKa = 3.85LEE602 pKa = 5.03KK603 pKa = 10.5ILKK606 pKa = 10.42DD607 pKa = 3.34SLDD610 pKa = 3.54PDD612 pKa = 3.71RR613 pKa = 11.84PQWGKK618 pKa = 10.81AIRR621 pKa = 11.84KK622 pKa = 9.16RR623 pKa = 11.84EE624 pKa = 3.87RR625 pKa = 11.84GKK627 pKa = 10.1IRR629 pKa = 11.84AVLGMDD635 pKa = 3.49VEE637 pKa = 5.31TYY639 pKa = 11.24LPMSYY644 pKa = 9.45IDD646 pKa = 6.28DD647 pKa = 4.91IIQPYY652 pKa = 10.11IIGNEE657 pKa = 3.96LSNLWEE663 pKa = 4.41TNFQRR668 pKa = 11.84HH669 pKa = 5.56AKK671 pKa = 10.3DD672 pKa = 2.96VDD674 pKa = 3.69RR675 pKa = 11.84LILSIEE681 pKa = 3.92KK682 pKa = 9.31TKK684 pKa = 10.52WFIPLDD690 pKa = 3.45QSGFDD695 pKa = 3.45HH696 pKa = 7.15QINNNMLLLTLVIIKK711 pKa = 9.84SWLLSRR717 pKa = 11.84VKK719 pKa = 10.96YY720 pKa = 10.51KK721 pKa = 11.11GDD723 pKa = 3.33VEE725 pKa = 4.25RR726 pKa = 11.84CIEE729 pKa = 4.2IIIKK733 pKa = 10.32RR734 pKa = 11.84LALTNSEE741 pKa = 4.41VNVDD745 pKa = 3.85DD746 pKa = 5.43KK747 pKa = 11.75INIPVEE753 pKa = 4.0RR754 pKa = 11.84GILSGWKK761 pKa = 7.04WTAFMDD767 pKa = 3.57TLINIGEE774 pKa = 4.36VNVALRR780 pKa = 11.84TCEE783 pKa = 3.91YY784 pKa = 10.62YY785 pKa = 11.23GLVCNYY791 pKa = 10.41KK792 pKa = 10.36LDD794 pKa = 4.0AQGDD798 pKa = 3.95DD799 pKa = 4.38DD800 pKa = 6.96DD801 pKa = 6.99IIIDD805 pKa = 4.15DD806 pKa = 4.76LLTGLVMCEE815 pKa = 3.49AYY817 pKa = 10.08RR818 pKa = 11.84YY819 pKa = 9.91CNLKK823 pKa = 10.23INPAKK828 pKa = 10.46FFIKK832 pKa = 10.74NNIDD836 pKa = 3.08EE837 pKa = 4.48YY838 pKa = 11.03LRR840 pKa = 11.84RR841 pKa = 11.84VYY843 pKa = 11.11VDD845 pKa = 4.0GVSLGYY851 pKa = 8.48PARR854 pKa = 11.84MFLTFFEE861 pKa = 5.1SNPIRR866 pKa = 11.84NVPLSEE872 pKa = 3.9MSEE875 pKa = 3.9LRR877 pKa = 11.84SVIKK881 pKa = 10.7NWMDD885 pKa = 4.77LIRR888 pKa = 11.84RR889 pKa = 11.84RR890 pKa = 11.84SLNNEE895 pKa = 3.47RR896 pKa = 11.84WVKK899 pKa = 10.02LVKK902 pKa = 10.67LMEE905 pKa = 4.75RR906 pKa = 11.84DD907 pKa = 2.89ILIRR911 pKa = 11.84VNKK914 pKa = 8.77NRR916 pKa = 11.84KK917 pKa = 8.77ISSLSLEE924 pKa = 4.57TILL927 pKa = 5.58
Molecular weight: 108.29 kDa
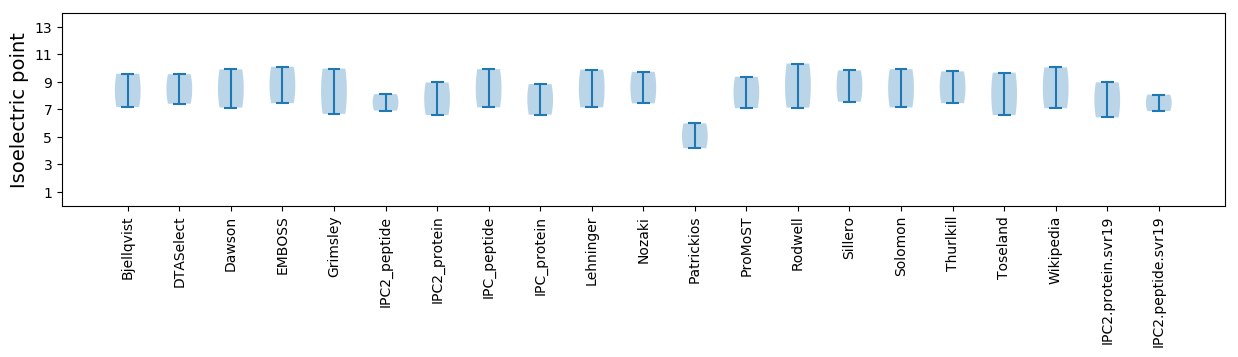
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E2RWR2|E2RWR2_9VIRU Putative coat protein OS=Rhizophagus sp. RF1 medium virus OX=758865 GN=CP PE=4 SV=1
MM1 pKa = 7.71SLRR4 pKa = 11.84LRR6 pKa = 11.84INTKK10 pKa = 9.77GLAVVIPSGMGKK22 pKa = 7.1TTLSNKK28 pKa = 9.55FPSLFLDD35 pKa = 3.73IDD37 pKa = 4.63DD38 pKa = 5.05IVWKK42 pKa = 9.71KK43 pKa = 10.32HH44 pKa = 5.94KK45 pKa = 10.24ILIGIINKK53 pKa = 9.32AFAWKK58 pKa = 10.18FGSDD62 pKa = 3.14LTKK65 pKa = 10.92NILRR69 pKa = 11.84RR70 pKa = 11.84KK71 pKa = 8.02WLWNDD76 pKa = 3.22TLSRR80 pKa = 11.84KK81 pKa = 9.35VLLCHH86 pKa = 7.01SPDD89 pKa = 3.54QLPSGVRR96 pKa = 11.84VLCILNSNSLNVNKK110 pKa = 10.09IKK112 pKa = 10.74KK113 pKa = 8.27EE114 pKa = 3.8RR115 pKa = 11.84KK116 pKa = 8.42ADD118 pKa = 3.5NVRR121 pKa = 11.84ISLQNRR127 pKa = 11.84HH128 pKa = 5.88SLWEE132 pKa = 3.89NYY134 pKa = 9.46RR135 pKa = 11.84DD136 pKa = 3.49EE137 pKa = 4.52EE138 pKa = 4.72NYY140 pKa = 10.53FEE142 pKa = 4.26VQSNKK147 pKa = 9.65VMLDD151 pKa = 3.33TIINCLIKK159 pKa = 10.92YY160 pKa = 10.39VDD162 pKa = 4.21FEE164 pKa = 4.57KK165 pKa = 11.12PNDD168 pKa = 3.61NTKK171 pKa = 10.9LLVPSKK177 pKa = 10.79DD178 pKa = 3.73YY179 pKa = 10.59IGKK182 pKa = 9.1FVRR185 pKa = 11.84KK186 pKa = 8.95EE187 pKa = 3.18RR188 pKa = 11.84YY189 pKa = 8.8LRR191 pKa = 11.84YY192 pKa = 9.19KK193 pKa = 9.98VKK195 pKa = 10.81YY196 pKa = 9.55DD197 pKa = 3.04ISEE200 pKa = 4.77PIVMSVRR207 pKa = 11.84EE208 pKa = 3.84YY209 pKa = 11.43MMIKK213 pKa = 10.25DD214 pKa = 4.01YY215 pKa = 10.89WRR217 pKa = 11.84DD218 pKa = 3.38LYY220 pKa = 10.16VHH222 pKa = 6.92RR223 pKa = 11.84KK224 pKa = 4.96WFKK227 pKa = 9.26IEE229 pKa = 4.54SYY231 pKa = 11.25GKK233 pKa = 9.73DD234 pKa = 3.29EE235 pKa = 4.2KK236 pKa = 11.14HH237 pKa = 6.58SLTDD241 pKa = 3.35EE242 pKa = 4.35SVNDD246 pKa = 3.72QIVTKK251 pKa = 10.29ILPLIDD257 pKa = 4.38KK258 pKa = 7.63EE259 pKa = 4.62TLFVLKK265 pKa = 10.1EE266 pKa = 3.97YY267 pKa = 10.39RR268 pKa = 11.84WDD270 pKa = 3.69RR271 pKa = 11.84MITNLMGIKK280 pKa = 10.24NSEE283 pKa = 4.17LVTNWMYY290 pKa = 10.87TPAYY294 pKa = 9.18LGGVGIQLDD303 pKa = 4.3GLRR306 pKa = 11.84KK307 pKa = 8.16PMSYY311 pKa = 8.39ITSSEE316 pKa = 4.27SIITPFVKK324 pKa = 10.08YY325 pKa = 9.55PKK327 pKa = 9.83SEE329 pKa = 3.97RR330 pKa = 11.84VEE332 pKa = 4.26NLTDD336 pKa = 3.98MILKK340 pKa = 9.97RR341 pKa = 11.84SKK343 pKa = 10.69FSVVDD348 pKa = 3.96NIQVYY353 pKa = 10.01EE354 pKa = 4.54GSQSTNKK361 pKa = 9.44IFKK364 pKa = 10.27SVGILRR370 pKa = 11.84FHH372 pKa = 7.07KK373 pKa = 10.95VIDD376 pKa = 4.27NINFRR381 pKa = 11.84KK382 pKa = 9.14FEE384 pKa = 4.01WMISGVDD391 pKa = 3.95FCKK394 pKa = 10.95SMIRR398 pKa = 11.84GVEE401 pKa = 4.0LPIKK405 pKa = 9.61MIRR408 pKa = 11.84PIFNSNIRR416 pKa = 11.84INQHH420 pKa = 5.77AQTIWINGLMRR431 pKa = 11.84AKK433 pKa = 10.61DD434 pKa = 3.6EE435 pKa = 4.87EE436 pKa = 4.32ITNEE440 pKa = 3.72ILSEE444 pKa = 4.07LFVNVSEE451 pKa = 5.62IIDD454 pKa = 3.46KK455 pKa = 10.61RR456 pKa = 11.84RR457 pKa = 11.84NFSKK461 pKa = 10.55IFWRR465 pKa = 11.84DD466 pKa = 3.23WLCGRR471 pKa = 11.84LEE473 pKa = 4.05YY474 pKa = 10.45QKK476 pKa = 10.33PINMWINDD484 pKa = 4.4NIVSTYY490 pKa = 10.33FIINLNRR497 pKa = 11.84LLSLNCNVDD506 pKa = 3.33YY507 pKa = 11.52GMLKK511 pKa = 10.51SMCKK515 pKa = 9.71IIEE518 pKa = 4.13KK519 pKa = 9.89TEE521 pKa = 3.87LNNIIRR527 pKa = 11.84YY528 pKa = 8.74LRR530 pKa = 11.84EE531 pKa = 3.63RR532 pKa = 11.84KK533 pKa = 7.96ITLMM537 pKa = 4.36
MM1 pKa = 7.71SLRR4 pKa = 11.84LRR6 pKa = 11.84INTKK10 pKa = 9.77GLAVVIPSGMGKK22 pKa = 7.1TTLSNKK28 pKa = 9.55FPSLFLDD35 pKa = 3.73IDD37 pKa = 4.63DD38 pKa = 5.05IVWKK42 pKa = 9.71KK43 pKa = 10.32HH44 pKa = 5.94KK45 pKa = 10.24ILIGIINKK53 pKa = 9.32AFAWKK58 pKa = 10.18FGSDD62 pKa = 3.14LTKK65 pKa = 10.92NILRR69 pKa = 11.84RR70 pKa = 11.84KK71 pKa = 8.02WLWNDD76 pKa = 3.22TLSRR80 pKa = 11.84KK81 pKa = 9.35VLLCHH86 pKa = 7.01SPDD89 pKa = 3.54QLPSGVRR96 pKa = 11.84VLCILNSNSLNVNKK110 pKa = 10.09IKK112 pKa = 10.74KK113 pKa = 8.27EE114 pKa = 3.8RR115 pKa = 11.84KK116 pKa = 8.42ADD118 pKa = 3.5NVRR121 pKa = 11.84ISLQNRR127 pKa = 11.84HH128 pKa = 5.88SLWEE132 pKa = 3.89NYY134 pKa = 9.46RR135 pKa = 11.84DD136 pKa = 3.49EE137 pKa = 4.52EE138 pKa = 4.72NYY140 pKa = 10.53FEE142 pKa = 4.26VQSNKK147 pKa = 9.65VMLDD151 pKa = 3.33TIINCLIKK159 pKa = 10.92YY160 pKa = 10.39VDD162 pKa = 4.21FEE164 pKa = 4.57KK165 pKa = 11.12PNDD168 pKa = 3.61NTKK171 pKa = 10.9LLVPSKK177 pKa = 10.79DD178 pKa = 3.73YY179 pKa = 10.59IGKK182 pKa = 9.1FVRR185 pKa = 11.84KK186 pKa = 8.95EE187 pKa = 3.18RR188 pKa = 11.84YY189 pKa = 8.8LRR191 pKa = 11.84YY192 pKa = 9.19KK193 pKa = 9.98VKK195 pKa = 10.81YY196 pKa = 9.55DD197 pKa = 3.04ISEE200 pKa = 4.77PIVMSVRR207 pKa = 11.84EE208 pKa = 3.84YY209 pKa = 11.43MMIKK213 pKa = 10.25DD214 pKa = 4.01YY215 pKa = 10.89WRR217 pKa = 11.84DD218 pKa = 3.38LYY220 pKa = 10.16VHH222 pKa = 6.92RR223 pKa = 11.84KK224 pKa = 4.96WFKK227 pKa = 9.26IEE229 pKa = 4.54SYY231 pKa = 11.25GKK233 pKa = 9.73DD234 pKa = 3.29EE235 pKa = 4.2KK236 pKa = 11.14HH237 pKa = 6.58SLTDD241 pKa = 3.35EE242 pKa = 4.35SVNDD246 pKa = 3.72QIVTKK251 pKa = 10.29ILPLIDD257 pKa = 4.38KK258 pKa = 7.63EE259 pKa = 4.62TLFVLKK265 pKa = 10.1EE266 pKa = 3.97YY267 pKa = 10.39RR268 pKa = 11.84WDD270 pKa = 3.69RR271 pKa = 11.84MITNLMGIKK280 pKa = 10.24NSEE283 pKa = 4.17LVTNWMYY290 pKa = 10.87TPAYY294 pKa = 9.18LGGVGIQLDD303 pKa = 4.3GLRR306 pKa = 11.84KK307 pKa = 8.16PMSYY311 pKa = 8.39ITSSEE316 pKa = 4.27SIITPFVKK324 pKa = 10.08YY325 pKa = 9.55PKK327 pKa = 9.83SEE329 pKa = 3.97RR330 pKa = 11.84VEE332 pKa = 4.26NLTDD336 pKa = 3.98MILKK340 pKa = 9.97RR341 pKa = 11.84SKK343 pKa = 10.69FSVVDD348 pKa = 3.96NIQVYY353 pKa = 10.01EE354 pKa = 4.54GSQSTNKK361 pKa = 9.44IFKK364 pKa = 10.27SVGILRR370 pKa = 11.84FHH372 pKa = 7.07KK373 pKa = 10.95VIDD376 pKa = 4.27NINFRR381 pKa = 11.84KK382 pKa = 9.14FEE384 pKa = 4.01WMISGVDD391 pKa = 3.95FCKK394 pKa = 10.95SMIRR398 pKa = 11.84GVEE401 pKa = 4.0LPIKK405 pKa = 9.61MIRR408 pKa = 11.84PIFNSNIRR416 pKa = 11.84INQHH420 pKa = 5.77AQTIWINGLMRR431 pKa = 11.84AKK433 pKa = 10.61DD434 pKa = 3.6EE435 pKa = 4.87EE436 pKa = 4.32ITNEE440 pKa = 3.72ILSEE444 pKa = 4.07LFVNVSEE451 pKa = 5.62IIDD454 pKa = 3.46KK455 pKa = 10.61RR456 pKa = 11.84RR457 pKa = 11.84NFSKK461 pKa = 10.55IFWRR465 pKa = 11.84DD466 pKa = 3.23WLCGRR471 pKa = 11.84LEE473 pKa = 4.05YY474 pKa = 10.45QKK476 pKa = 10.33PINMWINDD484 pKa = 4.4NIVSTYY490 pKa = 10.33FIINLNRR497 pKa = 11.84LLSLNCNVDD506 pKa = 3.33YY507 pKa = 11.52GMLKK511 pKa = 10.51SMCKK515 pKa = 9.71IIEE518 pKa = 4.13KK519 pKa = 9.89TEE521 pKa = 3.87LNNIIRR527 pKa = 11.84YY528 pKa = 8.74LRR530 pKa = 11.84EE531 pKa = 3.63RR532 pKa = 11.84KK533 pKa = 7.96ITLMM537 pKa = 4.36
Molecular weight: 63.59 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1464 |
537 |
927 |
732.0 |
85.94 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
2.391 ± 0.589 | 1.503 ± 0.108 |
7.104 ± 0.823 | 5.464 ± 0.066 |
4.167 ± 0.139 | 4.508 ± 0.425 |
1.434 ± 0.071 | 8.675 ± 1.254 |
8.06 ± 0.88 | 9.973 ± 0.157 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.801 ± 0.4 | 6.967 ± 0.463 |
3.415 ± 0.337 | 2.459 ± 0.324 |
6.352 ± 0.09 | 6.899 ± 0.197 |
3.962 ± 0.073 | 6.831 ± 0.17 |
2.254 ± 0.191 | 4.781 ± 0.472 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
