
Cangyuan orthoreovirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Resentoviricetes; Reovirales; Reoviridae; Spinareovirinae; Orthoreovirus; unclassified Orthoreovirus; Pteropine orthoreovirus
Average proteome isoelectric point is 6.31
Get precalculated fractions of proteins
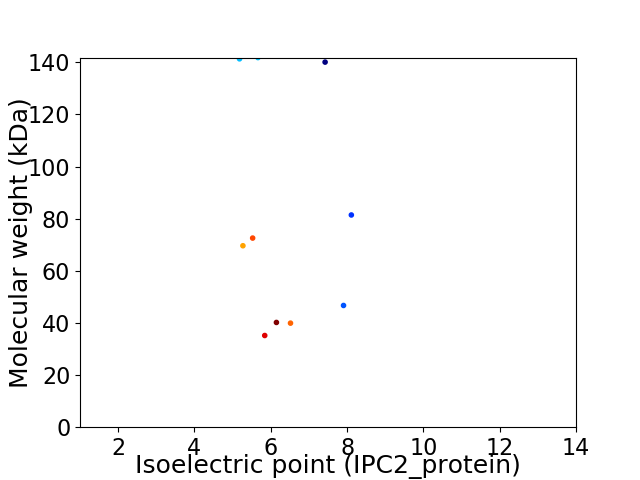
Virtual 2D-PAGE plot for 10 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A0A1ECY1|A0A0A1ECY1_9REOV Nonstructural protein OS=Cangyuan orthoreovirus OX=1388882 PE=4 SV=1
MM1 pKa = 7.06AQIRR5 pKa = 11.84GLRR8 pKa = 11.84LAEE11 pKa = 4.16SFNVPSKK18 pKa = 10.71SSSPSFITYY27 pKa = 10.63DD28 pKa = 3.5FLLSRR33 pKa = 11.84LQCASTPWTALTTSSGSDD51 pKa = 3.24VTVVRR56 pKa = 11.84LLFPLQGIQSPTLEE70 pKa = 4.31SPIPTTFSEE79 pKa = 4.25WKK81 pKa = 7.66TWMQEE86 pKa = 3.63RR87 pKa = 11.84LAVIYY92 pKa = 10.11EE93 pKa = 3.99DD94 pKa = 4.67LIRR97 pKa = 11.84RR98 pKa = 11.84YY99 pKa = 9.54PISEE103 pKa = 3.52FHH105 pKa = 6.15GRR107 pKa = 11.84HH108 pKa = 5.17VNPLICNTIVASFFSNSPYY127 pKa = 10.77SSLLDD132 pKa = 3.27HH133 pKa = 6.9VMLRR137 pKa = 11.84VSPAADD143 pKa = 2.91ILDD146 pKa = 4.01ANLLQRR152 pKa = 11.84NHH154 pKa = 6.25FWPLSNDD161 pKa = 3.82LLMMPAGKK169 pKa = 10.22KK170 pKa = 8.83YY171 pKa = 7.28WTFDD175 pKa = 3.27GYY177 pKa = 8.48QTSADD182 pKa = 3.84APCLFGKK189 pKa = 9.73NQTPYY194 pKa = 11.13ADD196 pKa = 3.63VIYY199 pKa = 10.72GRR201 pKa = 11.84DD202 pKa = 3.57LTSLLTFVDD211 pKa = 4.14SAPSQLTPLVHH222 pKa = 7.42LDD224 pKa = 3.35RR225 pKa = 11.84PSYY228 pKa = 10.09GPHH231 pKa = 5.64VLLPTEE237 pKa = 4.1NVIPAIATSSKK248 pKa = 11.18DD249 pKa = 3.38CAFWLLLEE257 pKa = 4.57SCLDD261 pKa = 3.47QLRR264 pKa = 11.84ANQSSSRR271 pKa = 11.84STPVTRR277 pKa = 11.84RR278 pKa = 11.84IHH280 pKa = 6.07SYY282 pKa = 10.25LVVRR286 pKa = 11.84SPYY289 pKa = 9.33FAFPEE294 pKa = 4.3SVEE297 pKa = 4.19YY298 pKa = 10.88ALLALNNASFCGSQLTVPVPNHH320 pKa = 5.64PTCSMVATLVSQLSDD335 pKa = 2.95SSKK338 pKa = 11.05AITVPSPKK346 pKa = 9.46PRR348 pKa = 11.84SFTLYY353 pKa = 10.82SNLKK357 pKa = 9.37FDD359 pKa = 4.62YY360 pKa = 9.32PQSLTPFISISPLTFTGTLGSASLLKK386 pKa = 9.16PTAASVRR393 pKa = 11.84WQPQFDD399 pKa = 4.7DD400 pKa = 4.15STSSPVDD407 pKa = 3.36LALNLMNSALTLPVTPDD424 pKa = 3.07YY425 pKa = 11.2GYY427 pKa = 11.33AWTGSPLFSYY437 pKa = 10.41AVSDD441 pKa = 3.51RR442 pKa = 11.84HH443 pKa = 6.27DD444 pKa = 3.48AAVPIDD450 pKa = 4.36LVPDD454 pKa = 5.05FPADD458 pKa = 3.63YY459 pKa = 10.44FSTEE463 pKa = 3.9QQDD466 pKa = 2.77ARR468 pKa = 11.84ARR470 pKa = 11.84FRR472 pKa = 11.84DD473 pKa = 3.4YY474 pKa = 11.31RR475 pKa = 11.84LIADD479 pKa = 4.44RR480 pKa = 11.84SLHH483 pKa = 6.83KK484 pKa = 10.38DD485 pKa = 3.21TANLACVSQQQDD497 pKa = 2.89ALGNRR502 pKa = 11.84FVFNGMSVAYY512 pKa = 8.74MGASGTHH519 pKa = 7.2PDD521 pKa = 4.33DD522 pKa = 3.8QPSVITPWLSGKK534 pKa = 9.52LASVFKK540 pKa = 10.19PSSIRR545 pKa = 11.84QFGWDD550 pKa = 3.3VTKK553 pKa = 10.96GVILDD558 pKa = 3.77VTHH561 pKa = 7.03SIASGDD567 pKa = 3.58FGFVYY572 pKa = 10.7SDD574 pKa = 3.33VDD576 pKa = 3.69QVQCNSDD583 pKa = 3.93DD584 pKa = 4.11LVSSTRR590 pKa = 11.84AFILQCQNLLTLVQVGGSLIVKK612 pKa = 9.79CNFPTSRR619 pKa = 11.84VLAWVYY625 pKa = 10.76TEE627 pKa = 4.14LSPWFNRR634 pKa = 11.84IIVMKK639 pKa = 10.01PLLSNNLEE647 pKa = 4.1IYY649 pKa = 10.6VGLLQKK655 pKa = 10.72LAITSPPFGPSSSVVVFLTKK675 pKa = 10.37QIRR678 pKa = 11.84RR679 pKa = 11.84YY680 pKa = 10.03SSLIEE685 pKa = 4.1ASVTLPDD692 pKa = 3.56RR693 pKa = 11.84GASVTLDD700 pKa = 3.27NSLTCLSLNFVNVTSVGSEE719 pKa = 3.79DD720 pKa = 4.16DD721 pKa = 4.57LRR723 pKa = 11.84ALACFSLLSSPSTIRR738 pKa = 11.84LSRR741 pKa = 11.84HH742 pKa = 5.48EE743 pKa = 4.3YY744 pKa = 9.77FDD746 pKa = 3.52SFRR749 pKa = 11.84TAVTSVVTPDD759 pKa = 4.1SRR761 pKa = 11.84RR762 pKa = 11.84LWSRR766 pKa = 11.84LAYY769 pKa = 9.71VPRR772 pKa = 11.84IFPSSLTVQSRR783 pKa = 11.84SIAYY787 pKa = 9.28SPPSLFKK794 pKa = 10.54TKK796 pKa = 10.15TSLWTLLSVYY806 pKa = 10.14YY807 pKa = 10.49DD808 pKa = 3.41WVLSGIPGRR817 pKa = 11.84PNLWMDD823 pKa = 4.41LGTGPEE829 pKa = 4.12CRR831 pKa = 11.84LLSKK835 pKa = 10.34ISEE838 pKa = 5.07DD839 pKa = 3.48IPVCMVDD846 pKa = 3.23VRR848 pKa = 11.84PSYY851 pKa = 11.44LPMNCWKK858 pKa = 8.6TQTDD862 pKa = 4.8YY863 pKa = 11.15IVKK866 pKa = 9.96DD867 pKa = 3.7YY868 pKa = 11.49TDD870 pKa = 3.35MQVILDD876 pKa = 3.9YY877 pKa = 11.54SPDD880 pKa = 3.64YY881 pKa = 11.2VSAILTLGAASFSSGVSLSSLVRR904 pKa = 11.84DD905 pKa = 4.37FLNSCKK911 pKa = 9.98QVGATKK917 pKa = 10.04IIFQLNSPADD927 pKa = 3.67TRR929 pKa = 11.84VDD931 pKa = 3.62SPHH934 pKa = 7.2RR935 pKa = 11.84EE936 pKa = 3.92LQIDD940 pKa = 3.65VSKK943 pKa = 11.33QMFFFPTLGRR953 pKa = 11.84KK954 pKa = 8.28EE955 pKa = 4.3PYY957 pKa = 10.36LPLTDD962 pKa = 4.77VIQLLSQTFPGATVEE977 pKa = 3.87IRR979 pKa = 11.84YY980 pKa = 9.5PEE982 pKa = 5.13DD983 pKa = 4.26DD984 pKa = 4.3YY985 pKa = 12.21SWLSNPLVNGLSIDD999 pKa = 3.37TDD1001 pKa = 4.57AISKK1005 pKa = 10.27LLVLSQFFPLFIVHH1019 pKa = 7.18ADD1021 pKa = 3.47VKK1023 pKa = 11.03SVTFSNVTSVGSEE1036 pKa = 3.71MSLTIHH1042 pKa = 6.59GFSSTSSYY1050 pKa = 11.63DD1051 pKa = 3.21VTLDD1055 pKa = 3.48GVSLLTISGAKK1066 pKa = 9.45VDD1068 pKa = 3.91SSLVSASLSVANDD1081 pKa = 3.52DD1082 pKa = 3.29AVLRR1086 pKa = 11.84FTPASAGILRR1096 pKa = 11.84VSQTAPVVLPIGSTVISAPDD1116 pKa = 3.44DD1117 pKa = 4.71AITVVWPSSLDD1128 pKa = 3.49YY1129 pKa = 11.37SDD1131 pKa = 5.5AGTNVSITCNSWFTLRR1147 pKa = 11.84LFAEE1151 pKa = 4.41RR1152 pKa = 11.84DD1153 pKa = 3.59GEE1155 pKa = 4.45VIRR1158 pKa = 11.84VSDD1161 pKa = 4.03DD1162 pKa = 3.32KK1163 pKa = 11.84YY1164 pKa = 10.99DD1165 pKa = 3.5IRR1167 pKa = 11.84PAGSNSWTLNVILDD1181 pKa = 3.96RR1182 pKa = 11.84SDD1184 pKa = 2.74AFYY1187 pKa = 11.06RR1188 pKa = 11.84FFLRR1192 pKa = 11.84DD1193 pKa = 3.34VQSAVPGQYY1202 pKa = 9.51IDD1204 pKa = 6.01FEE1206 pKa = 4.36LQQLSIHH1213 pKa = 5.68VWDD1216 pKa = 4.26PSKK1219 pKa = 10.84PSFLSPPYY1227 pKa = 10.41NGDD1230 pKa = 3.62FVISTPGGPVQLSRR1244 pKa = 11.84PFTQIPADD1252 pKa = 3.32WKK1254 pKa = 8.01TVNVSVSVISDD1265 pKa = 3.92LPSFLVPPSEE1275 pKa = 4.52YY1276 pKa = 10.71YY1277 pKa = 11.0GIVPVV1282 pKa = 3.94
MM1 pKa = 7.06AQIRR5 pKa = 11.84GLRR8 pKa = 11.84LAEE11 pKa = 4.16SFNVPSKK18 pKa = 10.71SSSPSFITYY27 pKa = 10.63DD28 pKa = 3.5FLLSRR33 pKa = 11.84LQCASTPWTALTTSSGSDD51 pKa = 3.24VTVVRR56 pKa = 11.84LLFPLQGIQSPTLEE70 pKa = 4.31SPIPTTFSEE79 pKa = 4.25WKK81 pKa = 7.66TWMQEE86 pKa = 3.63RR87 pKa = 11.84LAVIYY92 pKa = 10.11EE93 pKa = 3.99DD94 pKa = 4.67LIRR97 pKa = 11.84RR98 pKa = 11.84YY99 pKa = 9.54PISEE103 pKa = 3.52FHH105 pKa = 6.15GRR107 pKa = 11.84HH108 pKa = 5.17VNPLICNTIVASFFSNSPYY127 pKa = 10.77SSLLDD132 pKa = 3.27HH133 pKa = 6.9VMLRR137 pKa = 11.84VSPAADD143 pKa = 2.91ILDD146 pKa = 4.01ANLLQRR152 pKa = 11.84NHH154 pKa = 6.25FWPLSNDD161 pKa = 3.82LLMMPAGKK169 pKa = 10.22KK170 pKa = 8.83YY171 pKa = 7.28WTFDD175 pKa = 3.27GYY177 pKa = 8.48QTSADD182 pKa = 3.84APCLFGKK189 pKa = 9.73NQTPYY194 pKa = 11.13ADD196 pKa = 3.63VIYY199 pKa = 10.72GRR201 pKa = 11.84DD202 pKa = 3.57LTSLLTFVDD211 pKa = 4.14SAPSQLTPLVHH222 pKa = 7.42LDD224 pKa = 3.35RR225 pKa = 11.84PSYY228 pKa = 10.09GPHH231 pKa = 5.64VLLPTEE237 pKa = 4.1NVIPAIATSSKK248 pKa = 11.18DD249 pKa = 3.38CAFWLLLEE257 pKa = 4.57SCLDD261 pKa = 3.47QLRR264 pKa = 11.84ANQSSSRR271 pKa = 11.84STPVTRR277 pKa = 11.84RR278 pKa = 11.84IHH280 pKa = 6.07SYY282 pKa = 10.25LVVRR286 pKa = 11.84SPYY289 pKa = 9.33FAFPEE294 pKa = 4.3SVEE297 pKa = 4.19YY298 pKa = 10.88ALLALNNASFCGSQLTVPVPNHH320 pKa = 5.64PTCSMVATLVSQLSDD335 pKa = 2.95SSKK338 pKa = 11.05AITVPSPKK346 pKa = 9.46PRR348 pKa = 11.84SFTLYY353 pKa = 10.82SNLKK357 pKa = 9.37FDD359 pKa = 4.62YY360 pKa = 9.32PQSLTPFISISPLTFTGTLGSASLLKK386 pKa = 9.16PTAASVRR393 pKa = 11.84WQPQFDD399 pKa = 4.7DD400 pKa = 4.15STSSPVDD407 pKa = 3.36LALNLMNSALTLPVTPDD424 pKa = 3.07YY425 pKa = 11.2GYY427 pKa = 11.33AWTGSPLFSYY437 pKa = 10.41AVSDD441 pKa = 3.51RR442 pKa = 11.84HH443 pKa = 6.27DD444 pKa = 3.48AAVPIDD450 pKa = 4.36LVPDD454 pKa = 5.05FPADD458 pKa = 3.63YY459 pKa = 10.44FSTEE463 pKa = 3.9QQDD466 pKa = 2.77ARR468 pKa = 11.84ARR470 pKa = 11.84FRR472 pKa = 11.84DD473 pKa = 3.4YY474 pKa = 11.31RR475 pKa = 11.84LIADD479 pKa = 4.44RR480 pKa = 11.84SLHH483 pKa = 6.83KK484 pKa = 10.38DD485 pKa = 3.21TANLACVSQQQDD497 pKa = 2.89ALGNRR502 pKa = 11.84FVFNGMSVAYY512 pKa = 8.74MGASGTHH519 pKa = 7.2PDD521 pKa = 4.33DD522 pKa = 3.8QPSVITPWLSGKK534 pKa = 9.52LASVFKK540 pKa = 10.19PSSIRR545 pKa = 11.84QFGWDD550 pKa = 3.3VTKK553 pKa = 10.96GVILDD558 pKa = 3.77VTHH561 pKa = 7.03SIASGDD567 pKa = 3.58FGFVYY572 pKa = 10.7SDD574 pKa = 3.33VDD576 pKa = 3.69QVQCNSDD583 pKa = 3.93DD584 pKa = 4.11LVSSTRR590 pKa = 11.84AFILQCQNLLTLVQVGGSLIVKK612 pKa = 9.79CNFPTSRR619 pKa = 11.84VLAWVYY625 pKa = 10.76TEE627 pKa = 4.14LSPWFNRR634 pKa = 11.84IIVMKK639 pKa = 10.01PLLSNNLEE647 pKa = 4.1IYY649 pKa = 10.6VGLLQKK655 pKa = 10.72LAITSPPFGPSSSVVVFLTKK675 pKa = 10.37QIRR678 pKa = 11.84RR679 pKa = 11.84YY680 pKa = 10.03SSLIEE685 pKa = 4.1ASVTLPDD692 pKa = 3.56RR693 pKa = 11.84GASVTLDD700 pKa = 3.27NSLTCLSLNFVNVTSVGSEE719 pKa = 3.79DD720 pKa = 4.16DD721 pKa = 4.57LRR723 pKa = 11.84ALACFSLLSSPSTIRR738 pKa = 11.84LSRR741 pKa = 11.84HH742 pKa = 5.48EE743 pKa = 4.3YY744 pKa = 9.77FDD746 pKa = 3.52SFRR749 pKa = 11.84TAVTSVVTPDD759 pKa = 4.1SRR761 pKa = 11.84RR762 pKa = 11.84LWSRR766 pKa = 11.84LAYY769 pKa = 9.71VPRR772 pKa = 11.84IFPSSLTVQSRR783 pKa = 11.84SIAYY787 pKa = 9.28SPPSLFKK794 pKa = 10.54TKK796 pKa = 10.15TSLWTLLSVYY806 pKa = 10.14YY807 pKa = 10.49DD808 pKa = 3.41WVLSGIPGRR817 pKa = 11.84PNLWMDD823 pKa = 4.41LGTGPEE829 pKa = 4.12CRR831 pKa = 11.84LLSKK835 pKa = 10.34ISEE838 pKa = 5.07DD839 pKa = 3.48IPVCMVDD846 pKa = 3.23VRR848 pKa = 11.84PSYY851 pKa = 11.44LPMNCWKK858 pKa = 8.6TQTDD862 pKa = 4.8YY863 pKa = 11.15IVKK866 pKa = 9.96DD867 pKa = 3.7YY868 pKa = 11.49TDD870 pKa = 3.35MQVILDD876 pKa = 3.9YY877 pKa = 11.54SPDD880 pKa = 3.64YY881 pKa = 11.2VSAILTLGAASFSSGVSLSSLVRR904 pKa = 11.84DD905 pKa = 4.37FLNSCKK911 pKa = 9.98QVGATKK917 pKa = 10.04IIFQLNSPADD927 pKa = 3.67TRR929 pKa = 11.84VDD931 pKa = 3.62SPHH934 pKa = 7.2RR935 pKa = 11.84EE936 pKa = 3.92LQIDD940 pKa = 3.65VSKK943 pKa = 11.33QMFFFPTLGRR953 pKa = 11.84KK954 pKa = 8.28EE955 pKa = 4.3PYY957 pKa = 10.36LPLTDD962 pKa = 4.77VIQLLSQTFPGATVEE977 pKa = 3.87IRR979 pKa = 11.84YY980 pKa = 9.5PEE982 pKa = 5.13DD983 pKa = 4.26DD984 pKa = 4.3YY985 pKa = 12.21SWLSNPLVNGLSIDD999 pKa = 3.37TDD1001 pKa = 4.57AISKK1005 pKa = 10.27LLVLSQFFPLFIVHH1019 pKa = 7.18ADD1021 pKa = 3.47VKK1023 pKa = 11.03SVTFSNVTSVGSEE1036 pKa = 3.71MSLTIHH1042 pKa = 6.59GFSSTSSYY1050 pKa = 11.63DD1051 pKa = 3.21VTLDD1055 pKa = 3.48GVSLLTISGAKK1066 pKa = 9.45VDD1068 pKa = 3.91SSLVSASLSVANDD1081 pKa = 3.52DD1082 pKa = 3.29AVLRR1086 pKa = 11.84FTPASAGILRR1096 pKa = 11.84VSQTAPVVLPIGSTVISAPDD1116 pKa = 3.44DD1117 pKa = 4.71AITVVWPSSLDD1128 pKa = 3.49YY1129 pKa = 11.37SDD1131 pKa = 5.5AGTNVSITCNSWFTLRR1147 pKa = 11.84LFAEE1151 pKa = 4.41RR1152 pKa = 11.84DD1153 pKa = 3.59GEE1155 pKa = 4.45VIRR1158 pKa = 11.84VSDD1161 pKa = 4.03DD1162 pKa = 3.32KK1163 pKa = 11.84YY1164 pKa = 10.99DD1165 pKa = 3.5IRR1167 pKa = 11.84PAGSNSWTLNVILDD1181 pKa = 3.96RR1182 pKa = 11.84SDD1184 pKa = 2.74AFYY1187 pKa = 11.06RR1188 pKa = 11.84FFLRR1192 pKa = 11.84DD1193 pKa = 3.34VQSAVPGQYY1202 pKa = 9.51IDD1204 pKa = 6.01FEE1206 pKa = 4.36LQQLSIHH1213 pKa = 5.68VWDD1216 pKa = 4.26PSKK1219 pKa = 10.84PSFLSPPYY1227 pKa = 10.41NGDD1230 pKa = 3.62FVISTPGGPVQLSRR1244 pKa = 11.84PFTQIPADD1252 pKa = 3.32WKK1254 pKa = 8.01TVNVSVSVISDD1265 pKa = 3.92LPSFLVPPSEE1275 pKa = 4.52YY1276 pKa = 10.71YY1277 pKa = 11.0GIVPVV1282 pKa = 3.94
Molecular weight: 141.34 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0A1EAH6|A0A0A1EAH6_9REOV Nonstructural protein OS=Cangyuan orthoreovirus OX=1388882 PE=4 SV=1
MM1 pKa = 7.96AYY3 pKa = 10.33LALPVPEE10 pKa = 5.49GPDD13 pKa = 3.4SQSALISAVNGYY25 pKa = 9.95RR26 pKa = 11.84PDD28 pKa = 3.38GVAFDD33 pKa = 4.82DD34 pKa = 4.28VHH36 pKa = 8.29VLDD39 pKa = 4.19TKK41 pKa = 10.74RR42 pKa = 11.84VLRR45 pKa = 11.84QFDD48 pKa = 3.95LMTTGIRR55 pKa = 11.84ADD57 pKa = 3.89DD58 pKa = 3.74VAKK61 pKa = 10.85GLLLRR66 pKa = 11.84DD67 pKa = 3.44WRR69 pKa = 11.84RR70 pKa = 11.84QSVFVLLPQKK80 pKa = 9.82TSLLEE85 pKa = 4.09YY86 pKa = 10.73LLTAPKK92 pKa = 10.1TSLPDD97 pKa = 3.44GLTPQLLRR105 pKa = 11.84KK106 pKa = 9.98FKK108 pKa = 10.97AKK110 pKa = 10.18PHH112 pKa = 6.15DD113 pKa = 4.02FKK115 pKa = 11.58YY116 pKa = 10.98SDD118 pKa = 3.31WFSPLVTEE126 pKa = 4.93HH127 pKa = 6.69SSVVQSVRR135 pKa = 11.84YY136 pKa = 9.4LNSHH140 pKa = 6.08PVVFTTTVKK149 pKa = 10.95VIGAPVRR156 pKa = 11.84LFAPFKK162 pKa = 10.92YY163 pKa = 10.33FDD165 pKa = 3.54ISQGILSDD173 pKa = 4.76LMILKK178 pKa = 9.28MNHH181 pKa = 5.75QLPPLPAFRR190 pKa = 11.84VMVSMFPSSASGSCVLPPIDD210 pKa = 3.53EE211 pKa = 4.09WRR213 pKa = 11.84NSNTHH218 pKa = 5.77PVCLLLASMYY228 pKa = 10.57DD229 pKa = 3.39SSYY232 pKa = 11.07KK233 pKa = 9.63ATARR237 pKa = 11.84YY238 pKa = 9.54LDD240 pKa = 3.76RR241 pKa = 11.84SLITAMLCGQRR252 pKa = 11.84RR253 pKa = 11.84VKK255 pKa = 9.32QVKK258 pKa = 9.62YY259 pKa = 9.82SPLMARR265 pKa = 11.84AARR268 pKa = 11.84SVGLSVNVTNPTRR281 pKa = 11.84QINTSLIDD289 pKa = 3.34VHH291 pKa = 5.88TVVIDD296 pKa = 3.61VRR298 pKa = 11.84SSLDD302 pKa = 3.25NRR304 pKa = 11.84LTPTRR309 pKa = 11.84LRR311 pKa = 11.84FCGVPVALTAHH322 pKa = 6.36MGVAPSEE329 pKa = 4.14DD330 pKa = 3.03WLAVRR335 pKa = 11.84DD336 pKa = 3.97EE337 pKa = 4.41TGMFVDD343 pKa = 4.12WFLVLTLFSDD353 pKa = 4.73RR354 pKa = 11.84IKK356 pKa = 11.14GPTGQHH362 pKa = 4.6VCLNPLSTSVDD373 pKa = 3.14SVNFVRR379 pKa = 11.84VHH381 pKa = 6.36GYY383 pKa = 9.84VSQHH387 pKa = 4.69VQSLKK392 pKa = 7.85PWQYY396 pKa = 11.22GRR398 pKa = 11.84LSSFGVAMAKK408 pKa = 10.47GSFKK412 pKa = 10.63STMTRR417 pKa = 11.84FLTSLTIAGTRR428 pKa = 11.84LIFPNVIADD437 pKa = 3.58SDD439 pKa = 4.18DD440 pKa = 4.45PGDD443 pKa = 3.76SLEE446 pKa = 4.28PTFEE450 pKa = 4.1NQVLAEE456 pKa = 4.43LSSVDD461 pKa = 3.6ADD463 pKa = 3.49WEE465 pKa = 4.49RR466 pKa = 11.84KK467 pKa = 9.13VFTASGTVDD476 pKa = 3.27TSYY479 pKa = 11.35LSTVIFPIFLRR490 pKa = 11.84LFRR493 pKa = 11.84SEE495 pKa = 4.83LSPHH499 pKa = 6.02SRR501 pKa = 11.84TFYY504 pKa = 10.91DD505 pKa = 3.56EE506 pKa = 4.32RR507 pKa = 11.84ASSARR512 pKa = 11.84TLTFAHH518 pKa = 7.12ADD520 pKa = 3.6AEE522 pKa = 4.23FLDD525 pKa = 4.5AGWVDD530 pKa = 4.3RR531 pKa = 11.84IEE533 pKa = 4.03RR534 pKa = 11.84CYY536 pKa = 10.52IHH538 pKa = 7.24YY539 pKa = 10.61DD540 pKa = 3.3EE541 pKa = 5.12EE542 pKa = 5.53RR543 pKa = 11.84NVLLRR548 pKa = 11.84SSRR551 pKa = 11.84VGGSTFQLVLSRR563 pKa = 11.84CYY565 pKa = 11.28KK566 pKa = 10.01MIASPAPSEE575 pKa = 4.03PVSMLLKK582 pKa = 10.53ALVGGWLSAGPVLSSLSHH600 pKa = 5.69SASARR605 pKa = 11.84VLAWYY610 pKa = 9.72IDD612 pKa = 3.41DD613 pKa = 4.26HH614 pKa = 7.66HH615 pKa = 6.69WVDD618 pKa = 5.69HH619 pKa = 6.05GWCLCDD625 pKa = 3.6KK626 pKa = 9.66RR627 pKa = 11.84KK628 pKa = 9.87HH629 pKa = 4.3VTFSFMRR636 pKa = 11.84GHH638 pKa = 7.28PDD640 pKa = 3.23DD641 pKa = 5.52LAVLDD646 pKa = 4.08LQDD649 pKa = 3.14WSKK652 pKa = 11.13YY653 pKa = 8.75RR654 pKa = 11.84ATISVLTDD662 pKa = 3.57PLDD665 pKa = 4.13FGSSLRR671 pKa = 11.84VVAARR676 pKa = 11.84VYY678 pKa = 7.54WTSQKK683 pKa = 10.52PSVDD687 pKa = 3.38VFDD690 pKa = 4.47NRR692 pKa = 11.84ALLTPFQTYY701 pKa = 9.35HH702 pKa = 6.07VSLNCACPLGVRR714 pKa = 11.84FQVKK718 pKa = 9.94NVGLKK723 pKa = 10.22LATVSGAA730 pKa = 3.25
MM1 pKa = 7.96AYY3 pKa = 10.33LALPVPEE10 pKa = 5.49GPDD13 pKa = 3.4SQSALISAVNGYY25 pKa = 9.95RR26 pKa = 11.84PDD28 pKa = 3.38GVAFDD33 pKa = 4.82DD34 pKa = 4.28VHH36 pKa = 8.29VLDD39 pKa = 4.19TKK41 pKa = 10.74RR42 pKa = 11.84VLRR45 pKa = 11.84QFDD48 pKa = 3.95LMTTGIRR55 pKa = 11.84ADD57 pKa = 3.89DD58 pKa = 3.74VAKK61 pKa = 10.85GLLLRR66 pKa = 11.84DD67 pKa = 3.44WRR69 pKa = 11.84RR70 pKa = 11.84QSVFVLLPQKK80 pKa = 9.82TSLLEE85 pKa = 4.09YY86 pKa = 10.73LLTAPKK92 pKa = 10.1TSLPDD97 pKa = 3.44GLTPQLLRR105 pKa = 11.84KK106 pKa = 9.98FKK108 pKa = 10.97AKK110 pKa = 10.18PHH112 pKa = 6.15DD113 pKa = 4.02FKK115 pKa = 11.58YY116 pKa = 10.98SDD118 pKa = 3.31WFSPLVTEE126 pKa = 4.93HH127 pKa = 6.69SSVVQSVRR135 pKa = 11.84YY136 pKa = 9.4LNSHH140 pKa = 6.08PVVFTTTVKK149 pKa = 10.95VIGAPVRR156 pKa = 11.84LFAPFKK162 pKa = 10.92YY163 pKa = 10.33FDD165 pKa = 3.54ISQGILSDD173 pKa = 4.76LMILKK178 pKa = 9.28MNHH181 pKa = 5.75QLPPLPAFRR190 pKa = 11.84VMVSMFPSSASGSCVLPPIDD210 pKa = 3.53EE211 pKa = 4.09WRR213 pKa = 11.84NSNTHH218 pKa = 5.77PVCLLLASMYY228 pKa = 10.57DD229 pKa = 3.39SSYY232 pKa = 11.07KK233 pKa = 9.63ATARR237 pKa = 11.84YY238 pKa = 9.54LDD240 pKa = 3.76RR241 pKa = 11.84SLITAMLCGQRR252 pKa = 11.84RR253 pKa = 11.84VKK255 pKa = 9.32QVKK258 pKa = 9.62YY259 pKa = 9.82SPLMARR265 pKa = 11.84AARR268 pKa = 11.84SVGLSVNVTNPTRR281 pKa = 11.84QINTSLIDD289 pKa = 3.34VHH291 pKa = 5.88TVVIDD296 pKa = 3.61VRR298 pKa = 11.84SSLDD302 pKa = 3.25NRR304 pKa = 11.84LTPTRR309 pKa = 11.84LRR311 pKa = 11.84FCGVPVALTAHH322 pKa = 6.36MGVAPSEE329 pKa = 4.14DD330 pKa = 3.03WLAVRR335 pKa = 11.84DD336 pKa = 3.97EE337 pKa = 4.41TGMFVDD343 pKa = 4.12WFLVLTLFSDD353 pKa = 4.73RR354 pKa = 11.84IKK356 pKa = 11.14GPTGQHH362 pKa = 4.6VCLNPLSTSVDD373 pKa = 3.14SVNFVRR379 pKa = 11.84VHH381 pKa = 6.36GYY383 pKa = 9.84VSQHH387 pKa = 4.69VQSLKK392 pKa = 7.85PWQYY396 pKa = 11.22GRR398 pKa = 11.84LSSFGVAMAKK408 pKa = 10.47GSFKK412 pKa = 10.63STMTRR417 pKa = 11.84FLTSLTIAGTRR428 pKa = 11.84LIFPNVIADD437 pKa = 3.58SDD439 pKa = 4.18DD440 pKa = 4.45PGDD443 pKa = 3.76SLEE446 pKa = 4.28PTFEE450 pKa = 4.1NQVLAEE456 pKa = 4.43LSSVDD461 pKa = 3.6ADD463 pKa = 3.49WEE465 pKa = 4.49RR466 pKa = 11.84KK467 pKa = 9.13VFTASGTVDD476 pKa = 3.27TSYY479 pKa = 11.35LSTVIFPIFLRR490 pKa = 11.84LFRR493 pKa = 11.84SEE495 pKa = 4.83LSPHH499 pKa = 6.02SRR501 pKa = 11.84TFYY504 pKa = 10.91DD505 pKa = 3.56EE506 pKa = 4.32RR507 pKa = 11.84ASSARR512 pKa = 11.84TLTFAHH518 pKa = 7.12ADD520 pKa = 3.6AEE522 pKa = 4.23FLDD525 pKa = 4.5AGWVDD530 pKa = 4.3RR531 pKa = 11.84IEE533 pKa = 4.03RR534 pKa = 11.84CYY536 pKa = 10.52IHH538 pKa = 7.24YY539 pKa = 10.61DD540 pKa = 3.3EE541 pKa = 5.12EE542 pKa = 5.53RR543 pKa = 11.84NVLLRR548 pKa = 11.84SSRR551 pKa = 11.84VGGSTFQLVLSRR563 pKa = 11.84CYY565 pKa = 11.28KK566 pKa = 10.01MIASPAPSEE575 pKa = 4.03PVSMLLKK582 pKa = 10.53ALVGGWLSAGPVLSSLSHH600 pKa = 5.69SASARR605 pKa = 11.84VLAWYY610 pKa = 9.72IDD612 pKa = 3.41DD613 pKa = 4.26HH614 pKa = 7.66HH615 pKa = 6.69WVDD618 pKa = 5.69HH619 pKa = 6.05GWCLCDD625 pKa = 3.6KK626 pKa = 9.66RR627 pKa = 11.84KK628 pKa = 9.87HH629 pKa = 4.3VTFSFMRR636 pKa = 11.84GHH638 pKa = 7.28PDD640 pKa = 3.23DD641 pKa = 5.52LAVLDD646 pKa = 4.08LQDD649 pKa = 3.14WSKK652 pKa = 11.13YY653 pKa = 8.75RR654 pKa = 11.84ATISVLTDD662 pKa = 3.57PLDD665 pKa = 4.13FGSSLRR671 pKa = 11.84VVAARR676 pKa = 11.84VYY678 pKa = 7.54WTSQKK683 pKa = 10.52PSVDD687 pKa = 3.38VFDD690 pKa = 4.47NRR692 pKa = 11.84ALLTPFQTYY701 pKa = 9.35HH702 pKa = 6.07VSLNCACPLGVRR714 pKa = 11.84FQVKK718 pKa = 9.94NVGLKK723 pKa = 10.22LATVSGAA730 pKa = 3.25
Molecular weight: 81.43 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
7339 |
328 |
1290 |
733.9 |
80.89 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.094 ± 0.465 | 1.403 ± 0.073 |
5.832 ± 0.261 | 3.42 ± 0.458 |
3.992 ± 0.296 | 4.592 ± 0.261 |
1.908 ± 0.299 | 4.401 ± 0.356 |
3.311 ± 0.295 | 10.533 ± 0.387 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.739 ± 0.325 | 4.006 ± 0.464 |
6.295 ± 0.386 | 3.897 ± 0.264 |
5.409 ± 0.389 | 10.778 ± 0.714 |
7.113 ± 0.464 | 7.944 ± 0.331 |
1.676 ± 0.119 | 2.643 ± 0.314 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
