
Halomonas pacifica
Taxonomy: cellular organisms; Bacteria; Proteobacteria;
Average proteome isoelectric point is 6.2
Get precalculated fractions of proteins
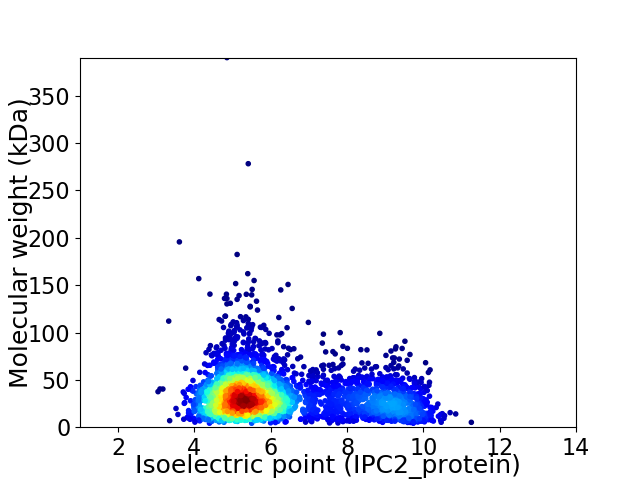
Virtual 2D-PAGE plot for 3562 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
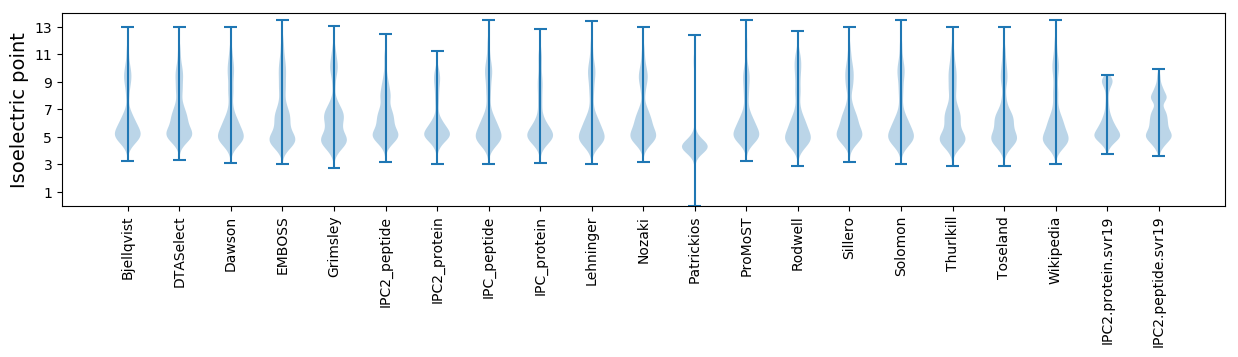
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A510XCT0|A0A510XCT0_9GAMM Transcriptional regulator OS=Halomonas pacifica OX=77098 GN=HPA02_06130 PE=4 SV=1
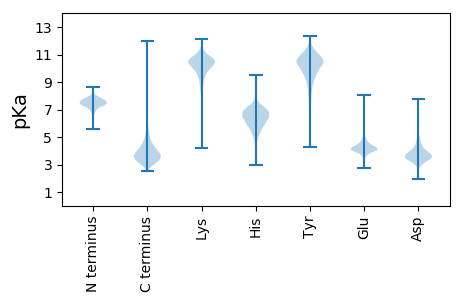
MM1 pKa = 7.71KK2 pKa = 9.06KK3 pKa = 8.27TLLATAIAGAMGLSAASAQAATVYY27 pKa = 10.67NQDD30 pKa = 3.01GTKK33 pKa = 10.43LDD35 pKa = 3.38IYY37 pKa = 11.46GNIQLAYY44 pKa = 9.51YY45 pKa = 10.5SVDD48 pKa = 3.03AGNNSVDD55 pKa = 3.55EE56 pKa = 4.38LADD59 pKa = 3.67NGSTFGFAAEE69 pKa = 4.25HH70 pKa = 6.26VINPGLTGYY79 pKa = 10.89LKK81 pKa = 11.21LEE83 pKa = 4.31FDD85 pKa = 4.44DD86 pKa = 6.14FKK88 pKa = 11.65ADD90 pKa = 4.11EE91 pKa = 4.59IKK93 pKa = 9.93TAGRR97 pKa = 11.84DD98 pKa = 3.5SGDD101 pKa = 3.12QAYY104 pKa = 10.71VGLRR108 pKa = 11.84GDD110 pKa = 3.76FGDD113 pKa = 4.55ARR115 pKa = 11.84LGSYY119 pKa = 10.07DD120 pKa = 4.35ALIDD124 pKa = 4.71DD125 pKa = 5.85WIQDD129 pKa = 4.02PITNNEE135 pKa = 3.83FFDD138 pKa = 4.31VNDD141 pKa = 3.68SSSQRR146 pKa = 11.84GVGGINEE153 pKa = 4.34TDD155 pKa = 2.97KK156 pKa = 10.67FTYY159 pKa = 9.41TSPSFGGFQFALGTQYY175 pKa = 10.91KK176 pKa = 10.56GEE178 pKa = 4.25AEE180 pKa = 4.28SEE182 pKa = 4.26NVTDD186 pKa = 4.18SGNASFFGGARR197 pKa = 11.84YY198 pKa = 7.43TVGAFSVAAVYY209 pKa = 10.97DD210 pKa = 3.78NLDD213 pKa = 3.65NYY215 pKa = 11.21ADD217 pKa = 4.25AYY219 pKa = 10.18QNNGQFGDD227 pKa = 3.77QYY229 pKa = 11.64GLTAQYY235 pKa = 10.42TIDD238 pKa = 3.9SLRR241 pKa = 11.84LAAKK245 pKa = 10.13VEE247 pKa = 4.17HH248 pKa = 6.96FDD250 pKa = 4.15GDD252 pKa = 3.98MANTDD257 pKa = 3.34ADD259 pKa = 4.61LYY261 pKa = 11.13ALSARR266 pKa = 11.84YY267 pKa = 9.47GYY269 pKa = 11.06GVGDD273 pKa = 3.27VYY275 pKa = 11.24GAYY278 pKa = 9.96QRR280 pKa = 11.84VDD282 pKa = 3.71DD283 pKa = 5.38DD284 pKa = 4.95LEE286 pKa = 6.03ADD288 pKa = 3.53EE289 pKa = 5.62RR290 pKa = 11.84DD291 pKa = 3.75EE292 pKa = 4.43FVVGATYY299 pKa = 10.98NLSDD303 pKa = 5.42AMYY306 pKa = 8.57TWAEE310 pKa = 3.89AAWYY314 pKa = 10.0DD315 pKa = 3.57NAQDD319 pKa = 3.52VGDD322 pKa = 4.14GVAVGVTYY330 pKa = 10.66LFF332 pKa = 4.38
MM1 pKa = 7.71KK2 pKa = 9.06KK3 pKa = 8.27TLLATAIAGAMGLSAASAQAATVYY27 pKa = 10.67NQDD30 pKa = 3.01GTKK33 pKa = 10.43LDD35 pKa = 3.38IYY37 pKa = 11.46GNIQLAYY44 pKa = 9.51YY45 pKa = 10.5SVDD48 pKa = 3.03AGNNSVDD55 pKa = 3.55EE56 pKa = 4.38LADD59 pKa = 3.67NGSTFGFAAEE69 pKa = 4.25HH70 pKa = 6.26VINPGLTGYY79 pKa = 10.89LKK81 pKa = 11.21LEE83 pKa = 4.31FDD85 pKa = 4.44DD86 pKa = 6.14FKK88 pKa = 11.65ADD90 pKa = 4.11EE91 pKa = 4.59IKK93 pKa = 9.93TAGRR97 pKa = 11.84DD98 pKa = 3.5SGDD101 pKa = 3.12QAYY104 pKa = 10.71VGLRR108 pKa = 11.84GDD110 pKa = 3.76FGDD113 pKa = 4.55ARR115 pKa = 11.84LGSYY119 pKa = 10.07DD120 pKa = 4.35ALIDD124 pKa = 4.71DD125 pKa = 5.85WIQDD129 pKa = 4.02PITNNEE135 pKa = 3.83FFDD138 pKa = 4.31VNDD141 pKa = 3.68SSSQRR146 pKa = 11.84GVGGINEE153 pKa = 4.34TDD155 pKa = 2.97KK156 pKa = 10.67FTYY159 pKa = 9.41TSPSFGGFQFALGTQYY175 pKa = 10.91KK176 pKa = 10.56GEE178 pKa = 4.25AEE180 pKa = 4.28SEE182 pKa = 4.26NVTDD186 pKa = 4.18SGNASFFGGARR197 pKa = 11.84YY198 pKa = 7.43TVGAFSVAAVYY209 pKa = 10.97DD210 pKa = 3.78NLDD213 pKa = 3.65NYY215 pKa = 11.21ADD217 pKa = 4.25AYY219 pKa = 10.18QNNGQFGDD227 pKa = 3.77QYY229 pKa = 11.64GLTAQYY235 pKa = 10.42TIDD238 pKa = 3.9SLRR241 pKa = 11.84LAAKK245 pKa = 10.13VEE247 pKa = 4.17HH248 pKa = 6.96FDD250 pKa = 4.15GDD252 pKa = 3.98MANTDD257 pKa = 3.34ADD259 pKa = 4.61LYY261 pKa = 11.13ALSARR266 pKa = 11.84YY267 pKa = 9.47GYY269 pKa = 11.06GVGDD273 pKa = 3.27VYY275 pKa = 11.24GAYY278 pKa = 9.96QRR280 pKa = 11.84VDD282 pKa = 3.71DD283 pKa = 5.38DD284 pKa = 4.95LEE286 pKa = 6.03ADD288 pKa = 3.53EE289 pKa = 5.62RR290 pKa = 11.84DD291 pKa = 3.75EE292 pKa = 4.43FVVGATYY299 pKa = 10.98NLSDD303 pKa = 5.42AMYY306 pKa = 8.57TWAEE310 pKa = 3.89AAWYY314 pKa = 10.0DD315 pKa = 3.57NAQDD319 pKa = 3.52VGDD322 pKa = 4.14GVAVGVTYY330 pKa = 10.66LFF332 pKa = 4.38
Molecular weight: 35.72 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A510X987|A0A510X987_9GAMM Outer membrane protein OS=Halomonas pacifica OX=77098 GN=HPA02_22460 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.34RR12 pKa = 11.84KK13 pKa = 9.1RR14 pKa = 11.84VHH16 pKa = 6.26GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.3NGRR28 pKa = 11.84AVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.93GRR39 pKa = 11.84KK40 pKa = 8.85RR41 pKa = 11.84LSAA44 pKa = 3.96
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.34RR12 pKa = 11.84KK13 pKa = 9.1RR14 pKa = 11.84VHH16 pKa = 6.26GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.3NGRR28 pKa = 11.84AVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.93GRR39 pKa = 11.84KK40 pKa = 8.85RR41 pKa = 11.84LSAA44 pKa = 3.96
Molecular weight: 5.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1139457 |
40 |
3557 |
319.9 |
34.97 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.334 ± 0.053 | 0.949 ± 0.014 |
5.436 ± 0.037 | 6.801 ± 0.046 |
3.334 ± 0.027 | 8.616 ± 0.041 |
2.37 ± 0.021 | 4.289 ± 0.031 |
2.267 ± 0.032 | 12.239 ± 0.065 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.279 ± 0.022 | 2.158 ± 0.023 |
5.178 ± 0.03 | 3.646 ± 0.029 |
7.87 ± 0.046 | 4.905 ± 0.023 |
4.606 ± 0.026 | 6.916 ± 0.038 |
1.504 ± 0.018 | 2.303 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
