
Catenovulum agarivorans DS-2
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Alteromonadales; Alteromonadaceae; Catenovulum; Catenovulum agarivorans
Average proteome isoelectric point is 6.22
Get precalculated fractions of proteins
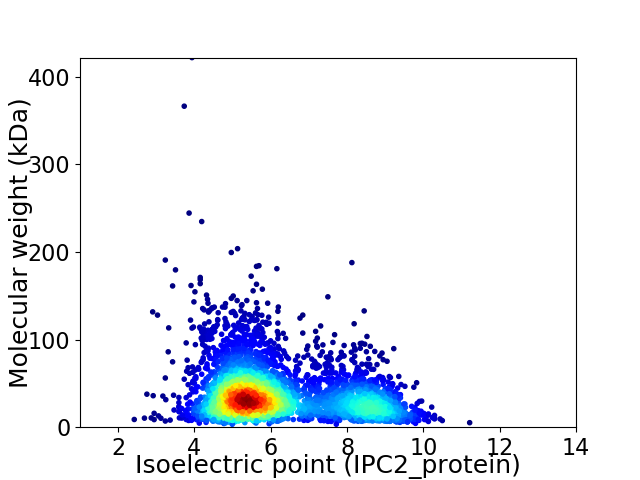
Virtual 2D-PAGE plot for 3835 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|W7QEY9|W7QEY9_9ALTE L-rhamnose mutarotase OS=Catenovulum agarivorans DS-2 OX=1328313 GN=rhaM PE=3 SV=1
MM1 pKa = 7.36SLSACKK7 pKa = 9.1DD8 pKa = 3.41TKK10 pKa = 11.14VDD12 pKa = 3.21IDD14 pKa = 3.98PPEE17 pKa = 4.52EE18 pKa = 3.98PTPAPSPTPTPTPTPTPTPTPANTAPTVDD47 pKa = 4.0MLDD50 pKa = 3.51SATVEE55 pKa = 3.83QGLSIEE61 pKa = 4.5LNWTISDD68 pKa = 4.08AEE70 pKa = 4.1QDD72 pKa = 3.67NLTCEE77 pKa = 3.91IDD79 pKa = 4.58AGDD82 pKa = 4.01SSNPIAITDD91 pKa = 3.61CAYY94 pKa = 9.76TGKK97 pKa = 7.64TTLTYY102 pKa = 10.31SEE104 pKa = 4.41SGQYY108 pKa = 9.44QIKK111 pKa = 8.21FTADD115 pKa = 3.15DD116 pKa = 4.61GEE118 pKa = 4.49FSSEE122 pKa = 3.59QTLNITISQPAPQFTLAASATAPAALYY149 pKa = 10.74LRR151 pKa = 11.84GFNDD155 pKa = 4.66DD156 pKa = 3.85WDD158 pKa = 4.11TTNPFTLHH166 pKa = 6.63DD167 pKa = 5.09GIYY170 pKa = 10.42VIEE173 pKa = 4.29FDD175 pKa = 3.96GTFAQQMVKK184 pKa = 10.18VASEE188 pKa = 3.84DD189 pKa = 3.24WSAVNCGGFALSANTAPVDD208 pKa = 4.01LTCGGNPDD216 pKa = 3.6NGSITFEE223 pKa = 3.65EE224 pKa = 4.47DD225 pKa = 2.5ALYY228 pKa = 10.49RR229 pKa = 11.84IGVSYY234 pKa = 10.92QDD236 pKa = 4.42DD237 pKa = 4.09GSAQMQMLKK246 pKa = 9.96IVDD249 pKa = 4.33PLPDD253 pKa = 3.57EE254 pKa = 5.03SGLGQAGTLNIHH266 pKa = 5.98YY267 pKa = 8.97LHH269 pKa = 6.91SSEE272 pKa = 5.7SYY274 pKa = 10.65SGWGLHH280 pKa = 5.69IWGDD284 pKa = 4.04AIDD287 pKa = 4.39PSIATTWQAPRR298 pKa = 11.84PYY300 pKa = 10.99AEE302 pKa = 5.34LKK304 pKa = 10.41DD305 pKa = 5.63DD306 pKa = 3.81YY307 pKa = 11.25AIWQVPIEE315 pKa = 4.49DD316 pKa = 4.25ANSAFNFIMHH326 pKa = 6.88FGDD329 pKa = 4.18VKK331 pKa = 10.9STDD334 pKa = 3.24SDD336 pKa = 3.91LSIVANSFGTDD347 pKa = 2.05IWIVQDD353 pKa = 3.24TATIYY358 pKa = 11.2NNEE361 pKa = 3.72TDD363 pKa = 4.57ARR365 pKa = 11.84AAYY368 pKa = 10.35NSLLQNLGNQSANLDD383 pKa = 3.83LSDD386 pKa = 3.62VTTTDD391 pKa = 2.91TSSALADD398 pKa = 3.45GWVDD402 pKa = 3.31SAQFAQIYY410 pKa = 8.62VRR412 pKa = 11.84AYY414 pKa = 9.96QDD416 pKa = 3.04SDD418 pKa = 3.47GDD420 pKa = 4.24GVGDD424 pKa = 3.61IQGLISRR431 pKa = 11.84LDD433 pKa = 3.54YY434 pKa = 11.48LKK436 pKa = 11.17DD437 pKa = 3.47NGIDD441 pKa = 4.34AIWLMPMMEE450 pKa = 4.92SSDD453 pKa = 3.77NDD455 pKa = 3.31HH456 pKa = 7.09GYY458 pKa = 8.7ATTDD462 pKa = 3.19YY463 pKa = 11.01RR464 pKa = 11.84KK465 pKa = 9.54IEE467 pKa = 4.06SDD469 pKa = 3.69YY470 pKa = 11.73GSLDD474 pKa = 3.49DD475 pKa = 6.43FKK477 pKa = 11.77ALLAEE482 pKa = 3.97AHH484 pKa = 6.07NRR486 pKa = 11.84DD487 pKa = 3.21MAIILDD493 pKa = 3.82YY494 pKa = 11.09VINHH498 pKa = 6.33SSNQNPLFADD508 pKa = 3.76AMSSANNDD516 pKa = 2.69KK517 pKa = 10.66RR518 pKa = 11.84DD519 pKa = 3.28WYY521 pKa = 10.02IIQADD526 pKa = 4.62KK527 pKa = 10.88PLGWNIWGGDD537 pKa = 3.59PWRR540 pKa = 11.84SSPGGAYY547 pKa = 9.62YY548 pKa = 10.83AAFADD553 pKa = 4.08SMPDD557 pKa = 3.36FDD559 pKa = 5.03LTNPDD564 pKa = 3.42VIAFHH569 pKa = 6.35QNNLKK574 pKa = 10.08FWLNMGVDD582 pKa = 4.3GFRR585 pKa = 11.84FDD587 pKa = 3.45AVGVLVEE594 pKa = 4.17NGKK597 pKa = 9.63DD598 pKa = 3.33ALEE601 pKa = 4.57DD602 pKa = 3.35QPEE605 pKa = 3.92NHH607 pKa = 6.79VIMKK611 pKa = 8.56TMKK614 pKa = 9.72DD615 pKa = 3.42TIEE618 pKa = 4.76SYY620 pKa = 10.31DD621 pKa = 3.28NRR623 pKa = 11.84YY624 pKa = 8.75IVCEE628 pKa = 4.18SPSGYY633 pKa = 10.79AAFANDD639 pKa = 3.74NSCGRR644 pKa = 11.84AFNFSAGHH652 pKa = 7.13AILGMVDD659 pKa = 3.48TGVVWDD665 pKa = 4.24EE666 pKa = 4.55LLTEE670 pKa = 4.62LAASNTDD677 pKa = 3.44QMPMILANHH686 pKa = 6.9DD687 pKa = 3.6FFAGDD692 pKa = 3.67RR693 pKa = 11.84VWNQLSGNVAQYY705 pKa = 11.24KK706 pKa = 9.77LAAATYY712 pKa = 10.02LLASSNPFTYY722 pKa = 10.32YY723 pKa = 10.84GEE725 pKa = 4.62EE726 pKa = 3.95IGLAAADD733 pKa = 4.17NLSGDD738 pKa = 3.32WSIRR742 pKa = 11.84TPMSWTDD749 pKa = 3.26DD750 pKa = 3.52TNNAGFTTGTPFRR763 pKa = 11.84DD764 pKa = 3.31LSSNVASNNVTAQLANADD782 pKa = 3.97SLLNYY787 pKa = 9.94YY788 pKa = 10.15RR789 pKa = 11.84DD790 pKa = 3.77LYY792 pKa = 9.54QVRR795 pKa = 11.84NDD797 pKa = 3.42HH798 pKa = 6.48SVIATGILNVQSSGGDD814 pKa = 3.17EE815 pKa = 4.07VLKK818 pKa = 10.6LVRR821 pKa = 11.84TGTGTEE827 pKa = 3.69AVILINYY834 pKa = 9.39SDD836 pKa = 4.11GDD838 pKa = 3.61EE839 pKa = 4.34SVTVDD844 pKa = 3.43VTHH847 pKa = 7.51DD848 pKa = 3.4NAVYY852 pKa = 10.0TDD854 pKa = 4.1EE855 pKa = 6.15LSDD858 pKa = 4.14LADD861 pKa = 4.26AGASSSGQLTVIVPAQSAAVFVLTVANN888 pKa = 4.03
MM1 pKa = 7.36SLSACKK7 pKa = 9.1DD8 pKa = 3.41TKK10 pKa = 11.14VDD12 pKa = 3.21IDD14 pKa = 3.98PPEE17 pKa = 4.52EE18 pKa = 3.98PTPAPSPTPTPTPTPTPTPTPANTAPTVDD47 pKa = 4.0MLDD50 pKa = 3.51SATVEE55 pKa = 3.83QGLSIEE61 pKa = 4.5LNWTISDD68 pKa = 4.08AEE70 pKa = 4.1QDD72 pKa = 3.67NLTCEE77 pKa = 3.91IDD79 pKa = 4.58AGDD82 pKa = 4.01SSNPIAITDD91 pKa = 3.61CAYY94 pKa = 9.76TGKK97 pKa = 7.64TTLTYY102 pKa = 10.31SEE104 pKa = 4.41SGQYY108 pKa = 9.44QIKK111 pKa = 8.21FTADD115 pKa = 3.15DD116 pKa = 4.61GEE118 pKa = 4.49FSSEE122 pKa = 3.59QTLNITISQPAPQFTLAASATAPAALYY149 pKa = 10.74LRR151 pKa = 11.84GFNDD155 pKa = 4.66DD156 pKa = 3.85WDD158 pKa = 4.11TTNPFTLHH166 pKa = 6.63DD167 pKa = 5.09GIYY170 pKa = 10.42VIEE173 pKa = 4.29FDD175 pKa = 3.96GTFAQQMVKK184 pKa = 10.18VASEE188 pKa = 3.84DD189 pKa = 3.24WSAVNCGGFALSANTAPVDD208 pKa = 4.01LTCGGNPDD216 pKa = 3.6NGSITFEE223 pKa = 3.65EE224 pKa = 4.47DD225 pKa = 2.5ALYY228 pKa = 10.49RR229 pKa = 11.84IGVSYY234 pKa = 10.92QDD236 pKa = 4.42DD237 pKa = 4.09GSAQMQMLKK246 pKa = 9.96IVDD249 pKa = 4.33PLPDD253 pKa = 3.57EE254 pKa = 5.03SGLGQAGTLNIHH266 pKa = 5.98YY267 pKa = 8.97LHH269 pKa = 6.91SSEE272 pKa = 5.7SYY274 pKa = 10.65SGWGLHH280 pKa = 5.69IWGDD284 pKa = 4.04AIDD287 pKa = 4.39PSIATTWQAPRR298 pKa = 11.84PYY300 pKa = 10.99AEE302 pKa = 5.34LKK304 pKa = 10.41DD305 pKa = 5.63DD306 pKa = 3.81YY307 pKa = 11.25AIWQVPIEE315 pKa = 4.49DD316 pKa = 4.25ANSAFNFIMHH326 pKa = 6.88FGDD329 pKa = 4.18VKK331 pKa = 10.9STDD334 pKa = 3.24SDD336 pKa = 3.91LSIVANSFGTDD347 pKa = 2.05IWIVQDD353 pKa = 3.24TATIYY358 pKa = 11.2NNEE361 pKa = 3.72TDD363 pKa = 4.57ARR365 pKa = 11.84AAYY368 pKa = 10.35NSLLQNLGNQSANLDD383 pKa = 3.83LSDD386 pKa = 3.62VTTTDD391 pKa = 2.91TSSALADD398 pKa = 3.45GWVDD402 pKa = 3.31SAQFAQIYY410 pKa = 8.62VRR412 pKa = 11.84AYY414 pKa = 9.96QDD416 pKa = 3.04SDD418 pKa = 3.47GDD420 pKa = 4.24GVGDD424 pKa = 3.61IQGLISRR431 pKa = 11.84LDD433 pKa = 3.54YY434 pKa = 11.48LKK436 pKa = 11.17DD437 pKa = 3.47NGIDD441 pKa = 4.34AIWLMPMMEE450 pKa = 4.92SSDD453 pKa = 3.77NDD455 pKa = 3.31HH456 pKa = 7.09GYY458 pKa = 8.7ATTDD462 pKa = 3.19YY463 pKa = 11.01RR464 pKa = 11.84KK465 pKa = 9.54IEE467 pKa = 4.06SDD469 pKa = 3.69YY470 pKa = 11.73GSLDD474 pKa = 3.49DD475 pKa = 6.43FKK477 pKa = 11.77ALLAEE482 pKa = 3.97AHH484 pKa = 6.07NRR486 pKa = 11.84DD487 pKa = 3.21MAIILDD493 pKa = 3.82YY494 pKa = 11.09VINHH498 pKa = 6.33SSNQNPLFADD508 pKa = 3.76AMSSANNDD516 pKa = 2.69KK517 pKa = 10.66RR518 pKa = 11.84DD519 pKa = 3.28WYY521 pKa = 10.02IIQADD526 pKa = 4.62KK527 pKa = 10.88PLGWNIWGGDD537 pKa = 3.59PWRR540 pKa = 11.84SSPGGAYY547 pKa = 9.62YY548 pKa = 10.83AAFADD553 pKa = 4.08SMPDD557 pKa = 3.36FDD559 pKa = 5.03LTNPDD564 pKa = 3.42VIAFHH569 pKa = 6.35QNNLKK574 pKa = 10.08FWLNMGVDD582 pKa = 4.3GFRR585 pKa = 11.84FDD587 pKa = 3.45AVGVLVEE594 pKa = 4.17NGKK597 pKa = 9.63DD598 pKa = 3.33ALEE601 pKa = 4.57DD602 pKa = 3.35QPEE605 pKa = 3.92NHH607 pKa = 6.79VIMKK611 pKa = 8.56TMKK614 pKa = 9.72DD615 pKa = 3.42TIEE618 pKa = 4.76SYY620 pKa = 10.31DD621 pKa = 3.28NRR623 pKa = 11.84YY624 pKa = 8.75IVCEE628 pKa = 4.18SPSGYY633 pKa = 10.79AAFANDD639 pKa = 3.74NSCGRR644 pKa = 11.84AFNFSAGHH652 pKa = 7.13AILGMVDD659 pKa = 3.48TGVVWDD665 pKa = 4.24EE666 pKa = 4.55LLTEE670 pKa = 4.62LAASNTDD677 pKa = 3.44QMPMILANHH686 pKa = 6.9DD687 pKa = 3.6FFAGDD692 pKa = 3.67RR693 pKa = 11.84VWNQLSGNVAQYY705 pKa = 11.24KK706 pKa = 9.77LAAATYY712 pKa = 10.02LLASSNPFTYY722 pKa = 10.32YY723 pKa = 10.84GEE725 pKa = 4.62EE726 pKa = 3.95IGLAAADD733 pKa = 4.17NLSGDD738 pKa = 3.32WSIRR742 pKa = 11.84TPMSWTDD749 pKa = 3.26DD750 pKa = 3.52TNNAGFTTGTPFRR763 pKa = 11.84DD764 pKa = 3.31LSSNVASNNVTAQLANADD782 pKa = 3.97SLLNYY787 pKa = 9.94YY788 pKa = 10.15RR789 pKa = 11.84DD790 pKa = 3.77LYY792 pKa = 9.54QVRR795 pKa = 11.84NDD797 pKa = 3.42HH798 pKa = 6.48SVIATGILNVQSSGGDD814 pKa = 3.17EE815 pKa = 4.07VLKK818 pKa = 10.6LVRR821 pKa = 11.84TGTGTEE827 pKa = 3.69AVILINYY834 pKa = 9.39SDD836 pKa = 4.11GDD838 pKa = 3.61EE839 pKa = 4.34SVTVDD844 pKa = 3.43VTHH847 pKa = 7.51DD848 pKa = 3.4NAVYY852 pKa = 10.0TDD854 pKa = 4.1EE855 pKa = 6.15LSDD858 pKa = 4.14LADD861 pKa = 4.26AGASSSGQLTVIVPAQSAAVFVLTVANN888 pKa = 4.03
Molecular weight: 96.17 kDa
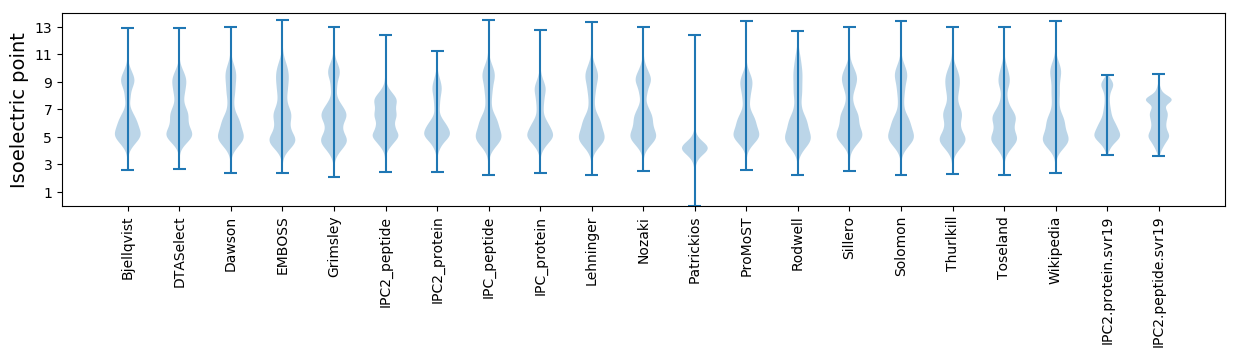
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|W7QVH6|W7QVH6_9ALTE Uncharacterized protein OS=Catenovulum agarivorans DS-2 OX=1328313 GN=DS2_02830 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 10.14RR12 pKa = 11.84KK13 pKa = 9.13RR14 pKa = 11.84SHH16 pKa = 6.17GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.4NGRR28 pKa = 11.84QVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.94GRR39 pKa = 11.84KK40 pKa = 8.67VLSAA44 pKa = 4.05
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 10.14RR12 pKa = 11.84KK13 pKa = 9.13RR14 pKa = 11.84SHH16 pKa = 6.17GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.4NGRR28 pKa = 11.84QVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.94GRR39 pKa = 11.84KK40 pKa = 8.67VLSAA44 pKa = 4.05
Molecular weight: 5.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1380918 |
28 |
3888 |
360.1 |
40.15 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.67 ± 0.038 | 0.977 ± 0.013 |
5.694 ± 0.037 | 5.869 ± 0.036 |
4.253 ± 0.022 | 6.316 ± 0.038 |
2.23 ± 0.021 | 6.427 ± 0.03 |
5.658 ± 0.04 | 9.978 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.198 ± 0.018 | 5.031 ± 0.035 |
3.726 ± 0.024 | 5.543 ± 0.046 |
3.955 ± 0.028 | 6.585 ± 0.04 |
5.449 ± 0.042 | 6.742 ± 0.037 |
1.303 ± 0.017 | 3.396 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
