
Bat Paramyxovirus Epo_spe/AR1/DRC/2009
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Paramyxoviridae; Rubulavirinae; Orthorubulavirus; Bat mumps orthorubulavirus
Average proteome isoelectric point is 6.48
Get precalculated fractions of proteins
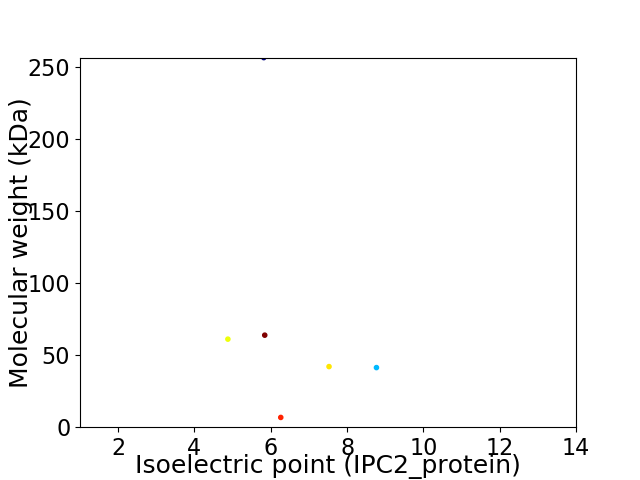
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|I0E095|I0E095_9MONO Phosphoprotein OS=Bat Paramyxovirus Epo_spe/AR1/DRC/2009 OX=1112597 GN=P PE=3 SV=1
MM1 pKa = 7.49SSVLKK6 pKa = 10.65AFEE9 pKa = 4.31RR10 pKa = 11.84FTIEE14 pKa = 5.13QEE16 pKa = 3.95LQDD19 pKa = 4.13RR20 pKa = 11.84GEE22 pKa = 4.35EE23 pKa = 4.24GAIPPEE29 pKa = 4.18TLKK32 pKa = 11.1SAVKK36 pKa = 10.23VFVINTPNPATRR48 pKa = 11.84YY49 pKa = 10.09QMLNFCLRR57 pKa = 11.84IICSQNARR65 pKa = 11.84ASHH68 pKa = 6.07RR69 pKa = 11.84VGALITLFSLPSAGMQNHH87 pKa = 6.05IRR89 pKa = 11.84LADD92 pKa = 4.27RR93 pKa = 11.84SPEE96 pKa = 3.9AQIEE100 pKa = 4.18RR101 pKa = 11.84CEE103 pKa = 4.02IDD105 pKa = 3.34GFEE108 pKa = 4.47PGTYY112 pKa = 10.03RR113 pKa = 11.84LIPNARR119 pKa = 11.84ANLTANEE126 pKa = 3.72IAAYY130 pKa = 10.25ALLADD135 pKa = 5.13DD136 pKa = 6.14LPPTINNGTPYY147 pKa = 10.34VHH149 pKa = 7.43ADD151 pKa = 3.72VEE153 pKa = 4.47GQPCDD158 pKa = 3.97EE159 pKa = 4.8IEE161 pKa = 4.09QFLDD165 pKa = 2.71RR166 pKa = 11.84CYY168 pKa = 11.21SVLIQAWVMVCKK180 pKa = 10.67CMTAYY185 pKa = 9.79DD186 pKa = 4.06QPAGSADD193 pKa = 3.45RR194 pKa = 11.84RR195 pKa = 11.84FAKK198 pKa = 10.22YY199 pKa = 8.47QQQGRR204 pKa = 11.84LEE206 pKa = 4.21ARR208 pKa = 11.84YY209 pKa = 8.28MLQPEE214 pKa = 4.38AQRR217 pKa = 11.84LIQAAIRR224 pKa = 11.84KK225 pKa = 8.85SLVVRR230 pKa = 11.84QYY232 pKa = 10.98LTFEE236 pKa = 4.12LQLARR241 pKa = 11.84RR242 pKa = 11.84QGLLSNRR249 pKa = 11.84YY250 pKa = 8.0YY251 pKa = 11.82AMVGDD256 pKa = 3.41IGKK259 pKa = 10.26YY260 pKa = 9.45IEE262 pKa = 4.47NSGLTAFFLTLKK274 pKa = 10.27YY275 pKa = 10.86ALGTKK280 pKa = 9.03WSPLSLAAFTGEE292 pKa = 4.05LTKK295 pKa = 10.79LRR297 pKa = 11.84SLMMLYY303 pKa = 10.24RR304 pKa = 11.84DD305 pKa = 3.69LGEE308 pKa = 3.91QARR311 pKa = 11.84YY312 pKa = 8.75LALLEE317 pKa = 4.89APQIMDD323 pKa = 4.55FAPGGYY329 pKa = 9.29PLIFSYY335 pKa = 11.38AMGVGTVLDD344 pKa = 3.74VQMRR348 pKa = 11.84NYY350 pKa = 9.87TYY352 pKa = 11.25ARR354 pKa = 11.84PFLNGYY360 pKa = 7.57YY361 pKa = 9.8FQIGVEE367 pKa = 4.14TARR370 pKa = 11.84RR371 pKa = 11.84QQGTVDD377 pKa = 3.5NRR379 pKa = 11.84VADD382 pKa = 4.32DD383 pKa = 4.52LGLTPEE389 pKa = 3.96QRR391 pKa = 11.84TEE393 pKa = 3.77VTQLVDD399 pKa = 3.29RR400 pKa = 11.84LARR403 pKa = 11.84GRR405 pKa = 11.84GAGIPGGPVNPFIPPAQQQQPAAVYY430 pKa = 10.97DD431 pKa = 3.97DD432 pKa = 3.99MPALEE437 pKa = 4.68EE438 pKa = 5.21SEE440 pKa = 6.1DD441 pKa = 5.18DD442 pKa = 5.07DD443 pKa = 7.39DD444 pKa = 7.14DD445 pKa = 6.69DD446 pKa = 6.29GGTRR450 pKa = 11.84FQNGAQAPIARR461 pKa = 11.84MGGQADD467 pKa = 4.15LRR469 pKa = 11.84AQHH472 pKa = 6.36AQDD475 pKa = 5.33PIQAQMFVPLYY486 pKa = 8.51PQAGNMPTQQSTQINQIGGADD507 pKa = 3.54QQDD510 pKa = 3.86LLRR513 pKa = 11.84YY514 pKa = 9.86NDD516 pKa = 5.85DD517 pKa = 3.36NDD519 pKa = 3.6QPQGARR525 pKa = 11.84GEE527 pKa = 4.3YY528 pKa = 10.84GSIFSNNPTQVPQSQVGDD546 pKa = 3.42WDD548 pKa = 3.74EE549 pKa = 3.83
MM1 pKa = 7.49SSVLKK6 pKa = 10.65AFEE9 pKa = 4.31RR10 pKa = 11.84FTIEE14 pKa = 5.13QEE16 pKa = 3.95LQDD19 pKa = 4.13RR20 pKa = 11.84GEE22 pKa = 4.35EE23 pKa = 4.24GAIPPEE29 pKa = 4.18TLKK32 pKa = 11.1SAVKK36 pKa = 10.23VFVINTPNPATRR48 pKa = 11.84YY49 pKa = 10.09QMLNFCLRR57 pKa = 11.84IICSQNARR65 pKa = 11.84ASHH68 pKa = 6.07RR69 pKa = 11.84VGALITLFSLPSAGMQNHH87 pKa = 6.05IRR89 pKa = 11.84LADD92 pKa = 4.27RR93 pKa = 11.84SPEE96 pKa = 3.9AQIEE100 pKa = 4.18RR101 pKa = 11.84CEE103 pKa = 4.02IDD105 pKa = 3.34GFEE108 pKa = 4.47PGTYY112 pKa = 10.03RR113 pKa = 11.84LIPNARR119 pKa = 11.84ANLTANEE126 pKa = 3.72IAAYY130 pKa = 10.25ALLADD135 pKa = 5.13DD136 pKa = 6.14LPPTINNGTPYY147 pKa = 10.34VHH149 pKa = 7.43ADD151 pKa = 3.72VEE153 pKa = 4.47GQPCDD158 pKa = 3.97EE159 pKa = 4.8IEE161 pKa = 4.09QFLDD165 pKa = 2.71RR166 pKa = 11.84CYY168 pKa = 11.21SVLIQAWVMVCKK180 pKa = 10.67CMTAYY185 pKa = 9.79DD186 pKa = 4.06QPAGSADD193 pKa = 3.45RR194 pKa = 11.84RR195 pKa = 11.84FAKK198 pKa = 10.22YY199 pKa = 8.47QQQGRR204 pKa = 11.84LEE206 pKa = 4.21ARR208 pKa = 11.84YY209 pKa = 8.28MLQPEE214 pKa = 4.38AQRR217 pKa = 11.84LIQAAIRR224 pKa = 11.84KK225 pKa = 8.85SLVVRR230 pKa = 11.84QYY232 pKa = 10.98LTFEE236 pKa = 4.12LQLARR241 pKa = 11.84RR242 pKa = 11.84QGLLSNRR249 pKa = 11.84YY250 pKa = 8.0YY251 pKa = 11.82AMVGDD256 pKa = 3.41IGKK259 pKa = 10.26YY260 pKa = 9.45IEE262 pKa = 4.47NSGLTAFFLTLKK274 pKa = 10.27YY275 pKa = 10.86ALGTKK280 pKa = 9.03WSPLSLAAFTGEE292 pKa = 4.05LTKK295 pKa = 10.79LRR297 pKa = 11.84SLMMLYY303 pKa = 10.24RR304 pKa = 11.84DD305 pKa = 3.69LGEE308 pKa = 3.91QARR311 pKa = 11.84YY312 pKa = 8.75LALLEE317 pKa = 4.89APQIMDD323 pKa = 4.55FAPGGYY329 pKa = 9.29PLIFSYY335 pKa = 11.38AMGVGTVLDD344 pKa = 3.74VQMRR348 pKa = 11.84NYY350 pKa = 9.87TYY352 pKa = 11.25ARR354 pKa = 11.84PFLNGYY360 pKa = 7.57YY361 pKa = 9.8FQIGVEE367 pKa = 4.14TARR370 pKa = 11.84RR371 pKa = 11.84QQGTVDD377 pKa = 3.5NRR379 pKa = 11.84VADD382 pKa = 4.32DD383 pKa = 4.52LGLTPEE389 pKa = 3.96QRR391 pKa = 11.84TEE393 pKa = 3.77VTQLVDD399 pKa = 3.29RR400 pKa = 11.84LARR403 pKa = 11.84GRR405 pKa = 11.84GAGIPGGPVNPFIPPAQQQQPAAVYY430 pKa = 10.97DD431 pKa = 3.97DD432 pKa = 3.99MPALEE437 pKa = 4.68EE438 pKa = 5.21SEE440 pKa = 6.1DD441 pKa = 5.18DD442 pKa = 5.07DD443 pKa = 7.39DD444 pKa = 7.14DD445 pKa = 6.69DD446 pKa = 6.29GGTRR450 pKa = 11.84FQNGAQAPIARR461 pKa = 11.84MGGQADD467 pKa = 4.15LRR469 pKa = 11.84AQHH472 pKa = 6.36AQDD475 pKa = 5.33PIQAQMFVPLYY486 pKa = 8.51PQAGNMPTQQSTQINQIGGADD507 pKa = 3.54QQDD510 pKa = 3.86LLRR513 pKa = 11.84YY514 pKa = 9.86NDD516 pKa = 5.85DD517 pKa = 3.36NDD519 pKa = 3.6QPQGARR525 pKa = 11.84GEE527 pKa = 4.3YY528 pKa = 10.84GSIFSNNPTQVPQSQVGDD546 pKa = 3.42WDD548 pKa = 3.74EE549 pKa = 3.83
Molecular weight: 61.15 kDa
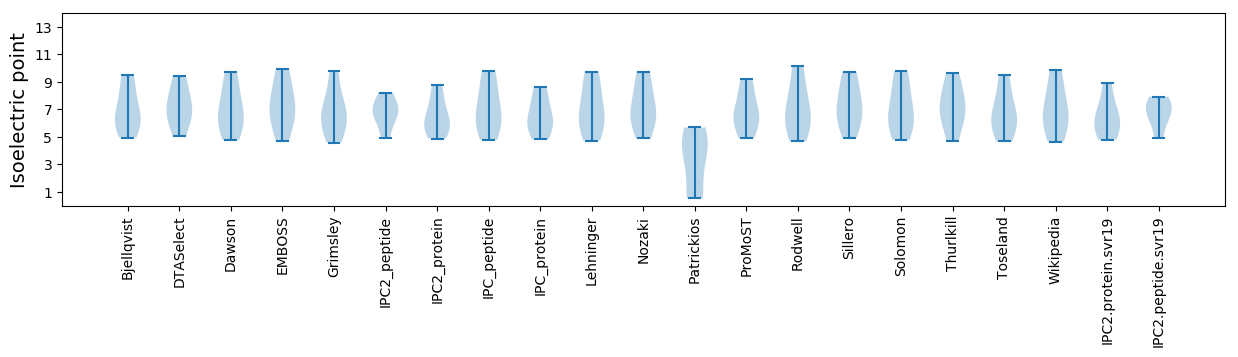
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|I0E097|I0E097_9MONO Small hydrophobic protein OS=Bat Paramyxovirus Epo_spe/AR1/DRC/2009 OX=1112597 GN=SH PE=3 SV=1
MM1 pKa = 7.74AGSQIKK7 pKa = 10.15IPLPKK12 pKa = 10.27PPDD15 pKa = 3.65SDD17 pKa = 3.86SQRR20 pKa = 11.84LNAFPIVMAQEE31 pKa = 4.3GKK33 pKa = 10.79GRR35 pKa = 11.84LLRR38 pKa = 11.84QIRR41 pKa = 11.84LRR43 pKa = 11.84KK44 pKa = 9.32LLSGDD49 pKa = 3.89PSDD52 pKa = 4.12HH53 pKa = 6.88QITFVNTYY61 pKa = 9.68GFIRR65 pKa = 11.84AAPEE69 pKa = 3.51TSEE72 pKa = 5.0FISEE76 pKa = 4.21SSPQKK81 pKa = 8.68ITPVVTACMLSFGAGPVLDD100 pKa = 5.91DD101 pKa = 3.97PQQMLKK107 pKa = 10.74ALDD110 pKa = 3.53QADD113 pKa = 2.94IRR115 pKa = 11.84VRR117 pKa = 11.84KK118 pKa = 6.83TASDD122 pKa = 3.62KK123 pKa = 10.74EE124 pKa = 4.17QVLFEE129 pKa = 4.55INRR132 pKa = 11.84IPNLFKK138 pKa = 10.5HH139 pKa = 5.81HH140 pKa = 6.43QISSDD145 pKa = 3.34HH146 pKa = 6.94LIQASSDD153 pKa = 3.64KK154 pKa = 10.67YY155 pKa = 10.17VKK157 pKa = 10.69SPAKK161 pKa = 9.34LTAGVNYY168 pKa = 9.75IYY170 pKa = 10.64CATFLSVTVCSASLKK185 pKa = 10.21FRR187 pKa = 11.84VARR190 pKa = 11.84PLLAARR196 pKa = 11.84SRR198 pKa = 11.84LVRR201 pKa = 11.84AVQMEE206 pKa = 3.97VLLRR210 pKa = 11.84VSCKK214 pKa = 9.82KK215 pKa = 10.35DD216 pKa = 3.26SPMAKK221 pKa = 10.19SMLTDD226 pKa = 4.09PDD228 pKa = 4.12GEE230 pKa = 4.48GCIASVWFHH239 pKa = 6.66LCNLCKK245 pKa = 10.4GRR247 pKa = 11.84NKK249 pKa = 10.08LRR251 pKa = 11.84SYY253 pKa = 10.8DD254 pKa = 3.72EE255 pKa = 4.62NYY257 pKa = 10.12FAAKK261 pKa = 8.66CKK263 pKa = 10.35KK264 pKa = 9.07MNLTVSVGDD273 pKa = 3.24MWGPTILVHH282 pKa = 7.2ASGHH286 pKa = 5.89IPTTAKK292 pKa = 10.4PFFNSRR298 pKa = 11.84GWVCHH303 pKa = 6.67PIHH306 pKa = 6.64QSSPSLAKK314 pKa = 9.57TLWSSGCEE322 pKa = 3.65IKK324 pKa = 10.75GASAILQGSDD334 pKa = 3.31YY335 pKa = 11.56ASLAKK340 pKa = 9.8TDD342 pKa = 4.63DD343 pKa = 3.84IIYY346 pKa = 10.72SKK348 pKa = 10.88IKK350 pKa = 9.61IDD352 pKa = 4.63KK353 pKa = 10.42DD354 pKa = 3.05AANYY358 pKa = 10.07KK359 pKa = 9.33GVSWNPFRR367 pKa = 11.84KK368 pKa = 9.38SASMNNLL375 pKa = 3.08
MM1 pKa = 7.74AGSQIKK7 pKa = 10.15IPLPKK12 pKa = 10.27PPDD15 pKa = 3.65SDD17 pKa = 3.86SQRR20 pKa = 11.84LNAFPIVMAQEE31 pKa = 4.3GKK33 pKa = 10.79GRR35 pKa = 11.84LLRR38 pKa = 11.84QIRR41 pKa = 11.84LRR43 pKa = 11.84KK44 pKa = 9.32LLSGDD49 pKa = 3.89PSDD52 pKa = 4.12HH53 pKa = 6.88QITFVNTYY61 pKa = 9.68GFIRR65 pKa = 11.84AAPEE69 pKa = 3.51TSEE72 pKa = 5.0FISEE76 pKa = 4.21SSPQKK81 pKa = 8.68ITPVVTACMLSFGAGPVLDD100 pKa = 5.91DD101 pKa = 3.97PQQMLKK107 pKa = 10.74ALDD110 pKa = 3.53QADD113 pKa = 2.94IRR115 pKa = 11.84VRR117 pKa = 11.84KK118 pKa = 6.83TASDD122 pKa = 3.62KK123 pKa = 10.74EE124 pKa = 4.17QVLFEE129 pKa = 4.55INRR132 pKa = 11.84IPNLFKK138 pKa = 10.5HH139 pKa = 5.81HH140 pKa = 6.43QISSDD145 pKa = 3.34HH146 pKa = 6.94LIQASSDD153 pKa = 3.64KK154 pKa = 10.67YY155 pKa = 10.17VKK157 pKa = 10.69SPAKK161 pKa = 9.34LTAGVNYY168 pKa = 9.75IYY170 pKa = 10.64CATFLSVTVCSASLKK185 pKa = 10.21FRR187 pKa = 11.84VARR190 pKa = 11.84PLLAARR196 pKa = 11.84SRR198 pKa = 11.84LVRR201 pKa = 11.84AVQMEE206 pKa = 3.97VLLRR210 pKa = 11.84VSCKK214 pKa = 9.82KK215 pKa = 10.35DD216 pKa = 3.26SPMAKK221 pKa = 10.19SMLTDD226 pKa = 4.09PDD228 pKa = 4.12GEE230 pKa = 4.48GCIASVWFHH239 pKa = 6.66LCNLCKK245 pKa = 10.4GRR247 pKa = 11.84NKK249 pKa = 10.08LRR251 pKa = 11.84SYY253 pKa = 10.8DD254 pKa = 3.72EE255 pKa = 4.62NYY257 pKa = 10.12FAAKK261 pKa = 8.66CKK263 pKa = 10.35KK264 pKa = 9.07MNLTVSVGDD273 pKa = 3.24MWGPTILVHH282 pKa = 7.2ASGHH286 pKa = 5.89IPTTAKK292 pKa = 10.4PFFNSRR298 pKa = 11.84GWVCHH303 pKa = 6.67PIHH306 pKa = 6.64QSSPSLAKK314 pKa = 9.57TLWSSGCEE322 pKa = 3.65IKK324 pKa = 10.75GASAILQGSDD334 pKa = 3.31YY335 pKa = 11.56ASLAKK340 pKa = 9.8TDD342 pKa = 4.63DD343 pKa = 3.84IIYY346 pKa = 10.72SKK348 pKa = 10.88IKK350 pKa = 9.61IDD352 pKa = 4.63KK353 pKa = 10.42DD354 pKa = 3.05AANYY358 pKa = 10.07KK359 pKa = 9.33GVSWNPFRR367 pKa = 11.84KK368 pKa = 9.38SASMNNLL375 pKa = 3.08
Molecular weight: 41.39 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4215 |
57 |
2261 |
702.5 |
78.66 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.216 ± 1.006 | 1.898 ± 0.289 |
5.219 ± 0.397 | 4.721 ± 0.561 |
3.369 ± 0.306 | 5.575 ± 0.813 |
1.708 ± 0.332 | 7.212 ± 0.51 |
4.555 ± 0.868 | 11.364 ± 1.274 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.752 ± 0.234 | 5.148 ± 0.54 |
5.362 ± 0.541 | 4.982 ± 0.963 |
4.958 ± 0.446 | 8.019 ± 0.807 |
6.358 ± 0.584 | 5.623 ± 0.368 |
1.186 ± 0.27 | 3.749 ± 0.481 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
