
Helminthosporium victoriae 145S virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Chrymotiviricetes; Ghabrivirales; Chrysoviridae; Chrysovirus
Average proteome isoelectric point is 5.99
Get precalculated fractions of proteins
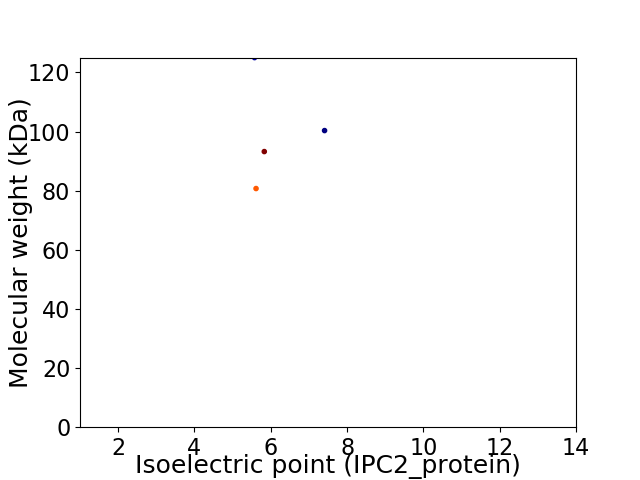
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
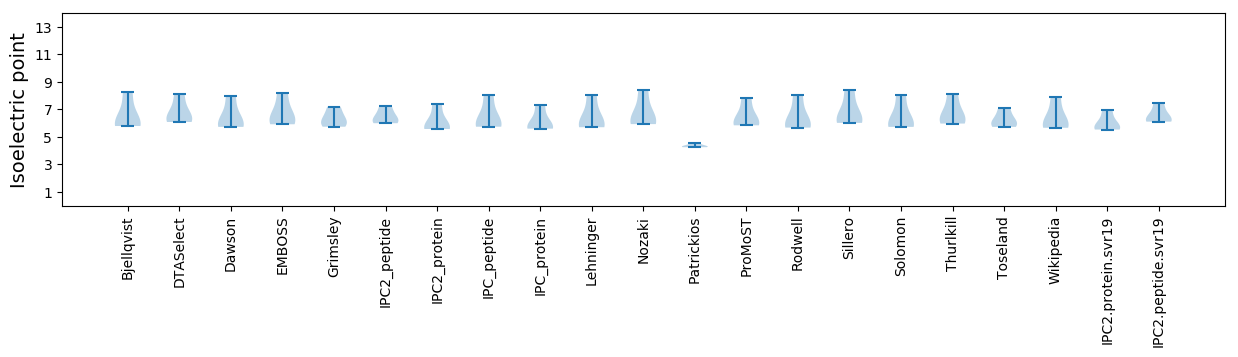
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q8JVB5|Q8JVB5_9VIRU Hv145SV-protein 3 OS=Helminthosporium victoriae 145S virus OX=164750 PE=2 SV=1
MM1 pKa = 7.32EE2 pKa = 6.42AIGHH6 pKa = 6.03LLKK9 pKa = 10.94GVAKK13 pKa = 10.48VNGDD17 pKa = 3.31RR18 pKa = 11.84RR19 pKa = 11.84VDD21 pKa = 3.37STMGSMVEE29 pKa = 3.83PTVNSRR35 pKa = 11.84GGMRR39 pKa = 11.84IEE41 pKa = 4.3RR42 pKa = 11.84NSKK45 pKa = 8.96VRR47 pKa = 11.84EE48 pKa = 4.03VNLARR53 pKa = 11.84AFRR56 pKa = 11.84KK57 pKa = 10.0SRR59 pKa = 11.84ITWKK63 pKa = 10.74LPAEE67 pKa = 4.21YY68 pKa = 9.54RR69 pKa = 11.84DD70 pKa = 3.86EE71 pKa = 4.07EE72 pKa = 4.99SMNIDD77 pKa = 4.39LRR79 pKa = 11.84PNLYY83 pKa = 9.28TVVMPAGCGKK93 pKa = 7.13TTIANEE99 pKa = 4.06FNCIDD104 pKa = 3.65VDD106 pKa = 4.03DD107 pKa = 4.74LAGVDD112 pKa = 4.23SRR114 pKa = 11.84AEE116 pKa = 3.6LMGWLKK122 pKa = 10.6EE123 pKa = 4.15ISDD126 pKa = 3.89GTASEE131 pKa = 3.95KK132 pKa = 10.97SEE134 pKa = 3.65WVRR137 pKa = 11.84IVNKK141 pKa = 10.45ALDD144 pKa = 3.45KK145 pKa = 10.66MVFEE149 pKa = 5.13EE150 pKa = 4.36PVVMLVHH157 pKa = 6.84DD158 pKa = 4.7HH159 pKa = 6.19LTAKK163 pKa = 10.56LVGSIRR169 pKa = 11.84AGTIMTPKK177 pKa = 10.42DD178 pKa = 3.34QVKK181 pKa = 10.37GMNKK185 pKa = 9.26NRR187 pKa = 11.84DD188 pKa = 3.82KK189 pKa = 10.88KK190 pKa = 9.69WNDD193 pKa = 2.53IFEE196 pKa = 4.65VTWAMAHH203 pKa = 6.74DD204 pKa = 4.55SNVKK208 pKa = 10.0RR209 pKa = 11.84KK210 pKa = 8.9WYY212 pKa = 9.02TPTTAEE218 pKa = 4.4CYY220 pKa = 9.63RR221 pKa = 11.84RR222 pKa = 11.84LARR225 pKa = 11.84VMANLHH231 pKa = 6.43CEE233 pKa = 4.06VPPPNVCFFDD243 pKa = 4.4AGMYY247 pKa = 10.82DD248 pKa = 3.34MEE250 pKa = 4.58LFKK253 pKa = 11.41GNEE256 pKa = 3.67NRR258 pKa = 11.84IEE260 pKa = 4.0EE261 pKa = 4.18LVEE264 pKa = 3.92LNEE267 pKa = 5.12AGLCPSLAIHH277 pKa = 6.62KK278 pKa = 9.64CCIKK282 pKa = 10.78NGMRR286 pKa = 11.84TEE288 pKa = 4.3MPWVSYY294 pKa = 11.06SKK296 pKa = 10.81LAGRR300 pKa = 11.84LANRR304 pKa = 11.84KK305 pKa = 8.7AAGVAGKK312 pKa = 10.49KK313 pKa = 10.05SGAIKK318 pKa = 10.52KK319 pKa = 10.23SLLWDD324 pKa = 3.41KK325 pKa = 11.11FNLGEE330 pKa = 4.7HH331 pKa = 6.66EE332 pKa = 4.55DD333 pKa = 3.6AVAIDD338 pKa = 3.62KK339 pKa = 11.05LLVKK343 pKa = 10.8SSDD346 pKa = 3.18AFSNCVVLWWKK357 pKa = 8.49TVMQEE362 pKa = 3.6YY363 pKa = 10.53DD364 pKa = 3.39IANHH368 pKa = 6.06IYY370 pKa = 11.07KK371 pKa = 9.73MILGVQEE378 pKa = 4.25KK379 pKa = 8.59EE380 pKa = 3.89WRR382 pKa = 11.84DD383 pKa = 3.41VLTDD387 pKa = 3.49IGQVLLTGAIGNVYY401 pKa = 10.14VSPEE405 pKa = 3.88SANVVMSARR414 pKa = 11.84AFSDD418 pKa = 3.62SEE420 pKa = 4.53GVTKK424 pKa = 10.71ACDD427 pKa = 3.21NWSLEE432 pKa = 4.12MNFEE436 pKa = 4.18EE437 pKa = 4.62TKK439 pKa = 10.51SDD441 pKa = 3.49VMVMIDD447 pKa = 4.31RR448 pKa = 11.84RR449 pKa = 11.84VLDD452 pKa = 4.18KK453 pKa = 11.16VSCGDD458 pKa = 3.55GEE460 pKa = 4.67YY461 pKa = 9.83MCAEE465 pKa = 4.44AEE467 pKa = 3.98WCRR470 pKa = 11.84WKK472 pKa = 10.96LDD474 pKa = 3.28DD475 pKa = 4.94CGLVRR480 pKa = 11.84GVVNITNMEE489 pKa = 4.05ADD491 pKa = 3.24DD492 pKa = 3.63QYY494 pKa = 11.76RR495 pKa = 11.84VVRR498 pKa = 11.84CIKK501 pKa = 10.24AIEE504 pKa = 4.5GGIQLEE510 pKa = 4.23PVIGSDD516 pKa = 3.47QYY518 pKa = 11.15LAEE521 pKa = 4.08MRR523 pKa = 11.84NARR526 pKa = 11.84EE527 pKa = 3.68VEE529 pKa = 4.2RR530 pKa = 11.84IVRR533 pKa = 11.84MSAYY537 pKa = 9.11KK538 pKa = 10.0VSAMGTILTTDD549 pKa = 4.25FEE551 pKa = 4.63ALPRR555 pKa = 11.84TRR557 pKa = 11.84CALEE561 pKa = 5.62AMFEE565 pKa = 4.13WTKK568 pKa = 10.21WLSGNYY574 pKa = 9.03EE575 pKa = 4.06EE576 pKa = 5.01YY577 pKa = 10.53LYY579 pKa = 11.16DD580 pKa = 3.42SVRR583 pKa = 11.84PTRR586 pKa = 11.84VIKK589 pKa = 8.98EE590 pKa = 3.56ICIPSSKK597 pKa = 10.91LEE599 pKa = 4.17MYY601 pKa = 10.72SGLTTMFMMGDD612 pKa = 3.22KK613 pKa = 10.09EE614 pKa = 4.23AAVVLAANAMRR625 pKa = 11.84NRR627 pKa = 11.84RR628 pKa = 11.84EE629 pKa = 3.62WTQDD633 pKa = 2.62VEE635 pKa = 4.36AYY637 pKa = 10.25HH638 pKa = 5.58MCEE641 pKa = 3.87VRR643 pKa = 11.84STEE646 pKa = 4.08SMRR649 pKa = 11.84LISKK653 pKa = 8.18SFRR656 pKa = 11.84EE657 pKa = 4.58AKK659 pKa = 10.15GHH661 pKa = 4.68IHH663 pKa = 7.33RR664 pKa = 11.84ICSSRR669 pKa = 11.84KK670 pKa = 9.39KK671 pKa = 9.75KK672 pKa = 10.78DD673 pKa = 3.21SVVWVEE679 pKa = 4.11DD680 pKa = 3.2STMEE684 pKa = 4.98DD685 pKa = 3.73MIDD688 pKa = 3.9EE689 pKa = 4.26ITVSIRR695 pKa = 11.84EE696 pKa = 4.07TMLCGDD702 pKa = 4.33FDD704 pKa = 4.03FTEE707 pKa = 4.44VSSASGG713 pKa = 3.09
MM1 pKa = 7.32EE2 pKa = 6.42AIGHH6 pKa = 6.03LLKK9 pKa = 10.94GVAKK13 pKa = 10.48VNGDD17 pKa = 3.31RR18 pKa = 11.84RR19 pKa = 11.84VDD21 pKa = 3.37STMGSMVEE29 pKa = 3.83PTVNSRR35 pKa = 11.84GGMRR39 pKa = 11.84IEE41 pKa = 4.3RR42 pKa = 11.84NSKK45 pKa = 8.96VRR47 pKa = 11.84EE48 pKa = 4.03VNLARR53 pKa = 11.84AFRR56 pKa = 11.84KK57 pKa = 10.0SRR59 pKa = 11.84ITWKK63 pKa = 10.74LPAEE67 pKa = 4.21YY68 pKa = 9.54RR69 pKa = 11.84DD70 pKa = 3.86EE71 pKa = 4.07EE72 pKa = 4.99SMNIDD77 pKa = 4.39LRR79 pKa = 11.84PNLYY83 pKa = 9.28TVVMPAGCGKK93 pKa = 7.13TTIANEE99 pKa = 4.06FNCIDD104 pKa = 3.65VDD106 pKa = 4.03DD107 pKa = 4.74LAGVDD112 pKa = 4.23SRR114 pKa = 11.84AEE116 pKa = 3.6LMGWLKK122 pKa = 10.6EE123 pKa = 4.15ISDD126 pKa = 3.89GTASEE131 pKa = 3.95KK132 pKa = 10.97SEE134 pKa = 3.65WVRR137 pKa = 11.84IVNKK141 pKa = 10.45ALDD144 pKa = 3.45KK145 pKa = 10.66MVFEE149 pKa = 5.13EE150 pKa = 4.36PVVMLVHH157 pKa = 6.84DD158 pKa = 4.7HH159 pKa = 6.19LTAKK163 pKa = 10.56LVGSIRR169 pKa = 11.84AGTIMTPKK177 pKa = 10.42DD178 pKa = 3.34QVKK181 pKa = 10.37GMNKK185 pKa = 9.26NRR187 pKa = 11.84DD188 pKa = 3.82KK189 pKa = 10.88KK190 pKa = 9.69WNDD193 pKa = 2.53IFEE196 pKa = 4.65VTWAMAHH203 pKa = 6.74DD204 pKa = 4.55SNVKK208 pKa = 10.0RR209 pKa = 11.84KK210 pKa = 8.9WYY212 pKa = 9.02TPTTAEE218 pKa = 4.4CYY220 pKa = 9.63RR221 pKa = 11.84RR222 pKa = 11.84LARR225 pKa = 11.84VMANLHH231 pKa = 6.43CEE233 pKa = 4.06VPPPNVCFFDD243 pKa = 4.4AGMYY247 pKa = 10.82DD248 pKa = 3.34MEE250 pKa = 4.58LFKK253 pKa = 11.41GNEE256 pKa = 3.67NRR258 pKa = 11.84IEE260 pKa = 4.0EE261 pKa = 4.18LVEE264 pKa = 3.92LNEE267 pKa = 5.12AGLCPSLAIHH277 pKa = 6.62KK278 pKa = 9.64CCIKK282 pKa = 10.78NGMRR286 pKa = 11.84TEE288 pKa = 4.3MPWVSYY294 pKa = 11.06SKK296 pKa = 10.81LAGRR300 pKa = 11.84LANRR304 pKa = 11.84KK305 pKa = 8.7AAGVAGKK312 pKa = 10.49KK313 pKa = 10.05SGAIKK318 pKa = 10.52KK319 pKa = 10.23SLLWDD324 pKa = 3.41KK325 pKa = 11.11FNLGEE330 pKa = 4.7HH331 pKa = 6.66EE332 pKa = 4.55DD333 pKa = 3.6AVAIDD338 pKa = 3.62KK339 pKa = 11.05LLVKK343 pKa = 10.8SSDD346 pKa = 3.18AFSNCVVLWWKK357 pKa = 8.49TVMQEE362 pKa = 3.6YY363 pKa = 10.53DD364 pKa = 3.39IANHH368 pKa = 6.06IYY370 pKa = 11.07KK371 pKa = 9.73MILGVQEE378 pKa = 4.25KK379 pKa = 8.59EE380 pKa = 3.89WRR382 pKa = 11.84DD383 pKa = 3.41VLTDD387 pKa = 3.49IGQVLLTGAIGNVYY401 pKa = 10.14VSPEE405 pKa = 3.88SANVVMSARR414 pKa = 11.84AFSDD418 pKa = 3.62SEE420 pKa = 4.53GVTKK424 pKa = 10.71ACDD427 pKa = 3.21NWSLEE432 pKa = 4.12MNFEE436 pKa = 4.18EE437 pKa = 4.62TKK439 pKa = 10.51SDD441 pKa = 3.49VMVMIDD447 pKa = 4.31RR448 pKa = 11.84RR449 pKa = 11.84VLDD452 pKa = 4.18KK453 pKa = 11.16VSCGDD458 pKa = 3.55GEE460 pKa = 4.67YY461 pKa = 9.83MCAEE465 pKa = 4.44AEE467 pKa = 3.98WCRR470 pKa = 11.84WKK472 pKa = 10.96LDD474 pKa = 3.28DD475 pKa = 4.94CGLVRR480 pKa = 11.84GVVNITNMEE489 pKa = 4.05ADD491 pKa = 3.24DD492 pKa = 3.63QYY494 pKa = 11.76RR495 pKa = 11.84VVRR498 pKa = 11.84CIKK501 pKa = 10.24AIEE504 pKa = 4.5GGIQLEE510 pKa = 4.23PVIGSDD516 pKa = 3.47QYY518 pKa = 11.15LAEE521 pKa = 4.08MRR523 pKa = 11.84NARR526 pKa = 11.84EE527 pKa = 3.68VEE529 pKa = 4.2RR530 pKa = 11.84IVRR533 pKa = 11.84MSAYY537 pKa = 9.11KK538 pKa = 10.0VSAMGTILTTDD549 pKa = 4.25FEE551 pKa = 4.63ALPRR555 pKa = 11.84TRR557 pKa = 11.84CALEE561 pKa = 5.62AMFEE565 pKa = 4.13WTKK568 pKa = 10.21WLSGNYY574 pKa = 9.03EE575 pKa = 4.06EE576 pKa = 5.01YY577 pKa = 10.53LYY579 pKa = 11.16DD580 pKa = 3.42SVRR583 pKa = 11.84PTRR586 pKa = 11.84VIKK589 pKa = 8.98EE590 pKa = 3.56ICIPSSKK597 pKa = 10.91LEE599 pKa = 4.17MYY601 pKa = 10.72SGLTTMFMMGDD612 pKa = 3.22KK613 pKa = 10.09EE614 pKa = 4.23AAVVLAANAMRR625 pKa = 11.84NRR627 pKa = 11.84RR628 pKa = 11.84EE629 pKa = 3.62WTQDD633 pKa = 2.62VEE635 pKa = 4.36AYY637 pKa = 10.25HH638 pKa = 5.58MCEE641 pKa = 3.87VRR643 pKa = 11.84STEE646 pKa = 4.08SMRR649 pKa = 11.84LISKK653 pKa = 8.18SFRR656 pKa = 11.84EE657 pKa = 4.58AKK659 pKa = 10.15GHH661 pKa = 4.68IHH663 pKa = 7.33RR664 pKa = 11.84ICSSRR669 pKa = 11.84KK670 pKa = 9.39KK671 pKa = 9.75KK672 pKa = 10.78DD673 pKa = 3.21SVVWVEE679 pKa = 4.11DD680 pKa = 3.2STMEE684 pKa = 4.98DD685 pKa = 3.73MIDD688 pKa = 3.9EE689 pKa = 4.26ITVSIRR695 pKa = 11.84EE696 pKa = 4.07TMLCGDD702 pKa = 4.33FDD704 pKa = 4.03FTEE707 pKa = 4.44VSSASGG713 pKa = 3.09
Molecular weight: 80.74 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q8JVB7|Q8JVB7_9VIRU RNA-directed RNA polymerase OS=Helminthosporium victoriae 145S virus OX=164750 PE=2 SV=1
MM1 pKa = 7.82ADD3 pKa = 3.33MFNAGKK9 pKa = 10.23YY10 pKa = 9.54DD11 pKa = 3.58RR12 pKa = 11.84RR13 pKa = 11.84QAIYY17 pKa = 8.28MAVRR21 pKa = 11.84AGAPAFKK28 pKa = 10.37KK29 pKa = 10.54AVEE32 pKa = 4.31TVQKK36 pKa = 10.64LEE38 pKa = 3.94HH39 pKa = 6.9WDD41 pKa = 3.49PTKK44 pKa = 10.92LPNKK48 pKa = 9.0MADD51 pKa = 3.61GKK53 pKa = 10.18PMLFKK58 pKa = 10.82GLQAVEE64 pKa = 4.82SYY66 pKa = 9.26MEE68 pKa = 4.61RR69 pKa = 11.84YY70 pKa = 9.06SASSLDD76 pKa = 3.64ALSEE80 pKa = 4.21EE81 pKa = 4.81KK82 pKa = 10.91NSFGYY87 pKa = 10.94GIFGDD92 pKa = 3.47IRR94 pKa = 11.84RR95 pKa = 11.84PAMLGANTVGIDD107 pKa = 3.44VTVEE111 pKa = 3.54WGSTEE116 pKa = 3.84VDD118 pKa = 2.93AEE120 pKa = 4.14MHH122 pKa = 5.98NGEE125 pKa = 4.27NRR127 pKa = 11.84KK128 pKa = 9.16MIVSTGTTTVRR139 pKa = 11.84NEE141 pKa = 3.84HH142 pKa = 5.92GKK144 pKa = 10.21DD145 pKa = 2.8RR146 pKa = 11.84AGISKK151 pKa = 8.7ATGWDD156 pKa = 3.07RR157 pKa = 11.84TEE159 pKa = 4.66CYY161 pKa = 10.77SMSPSDD167 pKa = 3.93VQTLCTLIDD176 pKa = 3.3TGRR179 pKa = 11.84AGFNKK184 pKa = 8.51YY185 pKa = 7.54TRR187 pKa = 11.84LVKK190 pKa = 10.66GMLVYY195 pKa = 10.83LDD197 pKa = 4.57LLNNGKK203 pKa = 8.46QAIKK207 pKa = 10.52KK208 pKa = 9.09RR209 pKa = 11.84IPNMISYY216 pKa = 10.0DD217 pKa = 3.75LKK219 pKa = 11.18SVLGSYY225 pKa = 7.02QHH227 pKa = 7.18RR228 pKa = 11.84DD229 pKa = 2.57RR230 pKa = 11.84SYY232 pKa = 10.49IYY234 pKa = 10.26CSNPDD239 pKa = 3.31SPEE242 pKa = 3.68YY243 pKa = 10.29RR244 pKa = 11.84VVLSLMSEE252 pKa = 4.91AYY254 pKa = 9.8PNDD257 pKa = 3.61DD258 pKa = 3.37MTCYY262 pKa = 10.4GVANIPADD270 pKa = 3.55GEE272 pKa = 4.67TNVIVVNGSAGSSVNYY288 pKa = 9.8HH289 pKa = 6.11VEE291 pKa = 4.0LTPQMVMASITQYY304 pKa = 9.91ATEE307 pKa = 4.34SGIADD312 pKa = 3.95EE313 pKa = 5.36LEE315 pKa = 4.23SALVCASSLYY325 pKa = 9.63HH326 pKa = 5.87NRR328 pKa = 11.84YY329 pKa = 7.95LARR332 pKa = 11.84VGLPRR337 pKa = 11.84VVSSIDD343 pKa = 3.75LIMPMFRR350 pKa = 11.84PANEE354 pKa = 3.89VRR356 pKa = 11.84CARR359 pKa = 11.84PSVAKK364 pKa = 10.04EE365 pKa = 3.42LAISIGKK372 pKa = 8.18LHH374 pKa = 5.65QMCMFLTIKK383 pKa = 10.51DD384 pKa = 3.47ILVAARR390 pKa = 11.84GSTKK394 pKa = 10.72AGFNYY399 pKa = 10.1SSVVEE404 pKa = 4.65SYY406 pKa = 9.49LTTQEE411 pKa = 4.14EE412 pKa = 5.27VVSVMNASVTPLRR425 pKa = 11.84LLEE428 pKa = 4.36MTPQMKK434 pKa = 9.5WMYY437 pKa = 10.39KK438 pKa = 9.28IDD440 pKa = 3.94AEE442 pKa = 4.55AMQDD446 pKa = 3.69LDD448 pKa = 3.71SLSIFEE454 pKa = 5.11VFWLCDD460 pKa = 3.27GGVASVRR467 pKa = 11.84NGGVMAFKK475 pKa = 10.63KK476 pKa = 10.46GVSDD480 pKa = 3.62MMTDD484 pKa = 3.39NPYY487 pKa = 10.77HH488 pKa = 7.17DD489 pKa = 4.22ILRR492 pKa = 11.84KK493 pKa = 9.49EE494 pKa = 3.97LAKK497 pKa = 11.04SNVVFDD503 pKa = 4.52FSKK506 pKa = 10.69LPKK509 pKa = 10.63GNFTIASRR517 pKa = 11.84YY518 pKa = 9.03IRR520 pKa = 11.84DD521 pKa = 3.6ANEE524 pKa = 3.62VVLPKK529 pKa = 10.51LEE531 pKa = 4.28YY532 pKa = 8.12VTEE535 pKa = 4.06RR536 pKa = 11.84VLIARR541 pKa = 11.84EE542 pKa = 3.91CDD544 pKa = 3.74YY545 pKa = 11.3NPHH548 pKa = 6.84DD549 pKa = 4.09RR550 pKa = 11.84VEE552 pKa = 4.49HH553 pKa = 6.44IINNRR558 pKa = 11.84TRR560 pKa = 11.84VTLSARR566 pKa = 11.84KK567 pKa = 8.57EE568 pKa = 3.93AMEE571 pKa = 4.21SGRR574 pKa = 11.84TKK576 pKa = 10.65VAWGDD581 pKa = 3.58IEE583 pKa = 4.51VEE585 pKa = 4.13RR586 pKa = 11.84PTGKK590 pKa = 10.69GSDD593 pKa = 3.5EE594 pKa = 4.4SEE596 pKa = 4.06SVLRR600 pKa = 11.84FGSSDD605 pKa = 2.91GRR607 pKa = 11.84QSPGIEE613 pKa = 3.8MFGLKK618 pKa = 9.97SEE620 pKa = 3.98ARR622 pKa = 11.84EE623 pKa = 3.77RR624 pKa = 11.84LEE626 pKa = 3.75KK627 pKa = 10.35RR628 pKa = 11.84RR629 pKa = 11.84SLVSPPPFRR638 pKa = 11.84QLSEE642 pKa = 4.13SASTRR647 pKa = 11.84RR648 pKa = 11.84AEE650 pKa = 3.97RR651 pKa = 11.84LSVSSHH657 pKa = 4.77GSRR660 pKa = 11.84RR661 pKa = 11.84SISVDD666 pKa = 3.28LEE668 pKa = 4.24SVRR671 pKa = 11.84SHH673 pKa = 6.78SVDD676 pKa = 3.18GDD678 pKa = 3.74DD679 pKa = 6.07DD680 pKa = 3.87KK681 pKa = 11.81TPTQSQNLRR690 pKa = 11.84KK691 pKa = 9.91RR692 pKa = 11.84FDD694 pKa = 3.59FSVLQKK700 pKa = 11.04AVDD703 pKa = 3.84EE704 pKa = 4.55KK705 pKa = 11.42KK706 pKa = 9.88MPGSYY711 pKa = 10.46EE712 pKa = 3.95STPEE716 pKa = 3.74KK717 pKa = 10.3TEE719 pKa = 3.69PTVTVEE725 pKa = 4.69KK726 pKa = 10.64IPGVKK731 pKa = 9.67SSMGVSEE738 pKa = 4.42EE739 pKa = 4.11VEE741 pKa = 3.72NDD743 pKa = 2.43KK744 pKa = 11.08RR745 pKa = 11.84AIYY748 pKa = 9.92KK749 pKa = 10.3ADD751 pKa = 3.72IIGSDD756 pKa = 4.14RR757 pKa = 11.84INGISAVNFHH767 pKa = 6.54KK768 pKa = 10.73LFRR771 pKa = 11.84EE772 pKa = 4.03RR773 pKa = 11.84FDD775 pKa = 3.75EE776 pKa = 4.39KK777 pKa = 10.74QVSPSVMARR786 pKa = 11.84LMAVLKK792 pKa = 10.76NVGVRR797 pKa = 11.84VNIAAMTADD806 pKa = 5.05EE807 pKa = 4.85INALMKK813 pKa = 10.24MRR815 pKa = 11.84DD816 pKa = 3.74GYY818 pKa = 11.1DD819 pKa = 2.77RR820 pKa = 11.84SYY822 pKa = 11.0RR823 pKa = 11.84SSEE826 pKa = 3.89SGHH829 pKa = 5.04NRR831 pKa = 11.84EE832 pKa = 3.98FRR834 pKa = 11.84EE835 pKa = 3.57PDD837 pKa = 3.13GRR839 pKa = 11.84VLTMEE844 pKa = 3.87INLRR848 pKa = 11.84RR849 pKa = 11.84MRR851 pKa = 11.84CDD853 pKa = 3.3RR854 pKa = 11.84DD855 pKa = 3.48KK856 pKa = 11.54KK857 pKa = 10.09PLRR860 pKa = 11.84EE861 pKa = 4.02ADD863 pKa = 3.67EE864 pKa = 4.54TGLIPDD870 pKa = 3.78HH871 pKa = 6.2MARR874 pKa = 11.84KK875 pKa = 9.77LGRR878 pKa = 11.84EE879 pKa = 4.0FFMAHH884 pKa = 6.66HH885 pKa = 6.47EE886 pKa = 4.01RR887 pKa = 11.84DD888 pKa = 3.55MILARR893 pKa = 11.84VVV895 pKa = 2.92
MM1 pKa = 7.82ADD3 pKa = 3.33MFNAGKK9 pKa = 10.23YY10 pKa = 9.54DD11 pKa = 3.58RR12 pKa = 11.84RR13 pKa = 11.84QAIYY17 pKa = 8.28MAVRR21 pKa = 11.84AGAPAFKK28 pKa = 10.37KK29 pKa = 10.54AVEE32 pKa = 4.31TVQKK36 pKa = 10.64LEE38 pKa = 3.94HH39 pKa = 6.9WDD41 pKa = 3.49PTKK44 pKa = 10.92LPNKK48 pKa = 9.0MADD51 pKa = 3.61GKK53 pKa = 10.18PMLFKK58 pKa = 10.82GLQAVEE64 pKa = 4.82SYY66 pKa = 9.26MEE68 pKa = 4.61RR69 pKa = 11.84YY70 pKa = 9.06SASSLDD76 pKa = 3.64ALSEE80 pKa = 4.21EE81 pKa = 4.81KK82 pKa = 10.91NSFGYY87 pKa = 10.94GIFGDD92 pKa = 3.47IRR94 pKa = 11.84RR95 pKa = 11.84PAMLGANTVGIDD107 pKa = 3.44VTVEE111 pKa = 3.54WGSTEE116 pKa = 3.84VDD118 pKa = 2.93AEE120 pKa = 4.14MHH122 pKa = 5.98NGEE125 pKa = 4.27NRR127 pKa = 11.84KK128 pKa = 9.16MIVSTGTTTVRR139 pKa = 11.84NEE141 pKa = 3.84HH142 pKa = 5.92GKK144 pKa = 10.21DD145 pKa = 2.8RR146 pKa = 11.84AGISKK151 pKa = 8.7ATGWDD156 pKa = 3.07RR157 pKa = 11.84TEE159 pKa = 4.66CYY161 pKa = 10.77SMSPSDD167 pKa = 3.93VQTLCTLIDD176 pKa = 3.3TGRR179 pKa = 11.84AGFNKK184 pKa = 8.51YY185 pKa = 7.54TRR187 pKa = 11.84LVKK190 pKa = 10.66GMLVYY195 pKa = 10.83LDD197 pKa = 4.57LLNNGKK203 pKa = 8.46QAIKK207 pKa = 10.52KK208 pKa = 9.09RR209 pKa = 11.84IPNMISYY216 pKa = 10.0DD217 pKa = 3.75LKK219 pKa = 11.18SVLGSYY225 pKa = 7.02QHH227 pKa = 7.18RR228 pKa = 11.84DD229 pKa = 2.57RR230 pKa = 11.84SYY232 pKa = 10.49IYY234 pKa = 10.26CSNPDD239 pKa = 3.31SPEE242 pKa = 3.68YY243 pKa = 10.29RR244 pKa = 11.84VVLSLMSEE252 pKa = 4.91AYY254 pKa = 9.8PNDD257 pKa = 3.61DD258 pKa = 3.37MTCYY262 pKa = 10.4GVANIPADD270 pKa = 3.55GEE272 pKa = 4.67TNVIVVNGSAGSSVNYY288 pKa = 9.8HH289 pKa = 6.11VEE291 pKa = 4.0LTPQMVMASITQYY304 pKa = 9.91ATEE307 pKa = 4.34SGIADD312 pKa = 3.95EE313 pKa = 5.36LEE315 pKa = 4.23SALVCASSLYY325 pKa = 9.63HH326 pKa = 5.87NRR328 pKa = 11.84YY329 pKa = 7.95LARR332 pKa = 11.84VGLPRR337 pKa = 11.84VVSSIDD343 pKa = 3.75LIMPMFRR350 pKa = 11.84PANEE354 pKa = 3.89VRR356 pKa = 11.84CARR359 pKa = 11.84PSVAKK364 pKa = 10.04EE365 pKa = 3.42LAISIGKK372 pKa = 8.18LHH374 pKa = 5.65QMCMFLTIKK383 pKa = 10.51DD384 pKa = 3.47ILVAARR390 pKa = 11.84GSTKK394 pKa = 10.72AGFNYY399 pKa = 10.1SSVVEE404 pKa = 4.65SYY406 pKa = 9.49LTTQEE411 pKa = 4.14EE412 pKa = 5.27VVSVMNASVTPLRR425 pKa = 11.84LLEE428 pKa = 4.36MTPQMKK434 pKa = 9.5WMYY437 pKa = 10.39KK438 pKa = 9.28IDD440 pKa = 3.94AEE442 pKa = 4.55AMQDD446 pKa = 3.69LDD448 pKa = 3.71SLSIFEE454 pKa = 5.11VFWLCDD460 pKa = 3.27GGVASVRR467 pKa = 11.84NGGVMAFKK475 pKa = 10.63KK476 pKa = 10.46GVSDD480 pKa = 3.62MMTDD484 pKa = 3.39NPYY487 pKa = 10.77HH488 pKa = 7.17DD489 pKa = 4.22ILRR492 pKa = 11.84KK493 pKa = 9.49EE494 pKa = 3.97LAKK497 pKa = 11.04SNVVFDD503 pKa = 4.52FSKK506 pKa = 10.69LPKK509 pKa = 10.63GNFTIASRR517 pKa = 11.84YY518 pKa = 9.03IRR520 pKa = 11.84DD521 pKa = 3.6ANEE524 pKa = 3.62VVLPKK529 pKa = 10.51LEE531 pKa = 4.28YY532 pKa = 8.12VTEE535 pKa = 4.06RR536 pKa = 11.84VLIARR541 pKa = 11.84EE542 pKa = 3.91CDD544 pKa = 3.74YY545 pKa = 11.3NPHH548 pKa = 6.84DD549 pKa = 4.09RR550 pKa = 11.84VEE552 pKa = 4.49HH553 pKa = 6.44IINNRR558 pKa = 11.84TRR560 pKa = 11.84VTLSARR566 pKa = 11.84KK567 pKa = 8.57EE568 pKa = 3.93AMEE571 pKa = 4.21SGRR574 pKa = 11.84TKK576 pKa = 10.65VAWGDD581 pKa = 3.58IEE583 pKa = 4.51VEE585 pKa = 4.13RR586 pKa = 11.84PTGKK590 pKa = 10.69GSDD593 pKa = 3.5EE594 pKa = 4.4SEE596 pKa = 4.06SVLRR600 pKa = 11.84FGSSDD605 pKa = 2.91GRR607 pKa = 11.84QSPGIEE613 pKa = 3.8MFGLKK618 pKa = 9.97SEE620 pKa = 3.98ARR622 pKa = 11.84EE623 pKa = 3.77RR624 pKa = 11.84LEE626 pKa = 3.75KK627 pKa = 10.35RR628 pKa = 11.84RR629 pKa = 11.84SLVSPPPFRR638 pKa = 11.84QLSEE642 pKa = 4.13SASTRR647 pKa = 11.84RR648 pKa = 11.84AEE650 pKa = 3.97RR651 pKa = 11.84LSVSSHH657 pKa = 4.77GSRR660 pKa = 11.84RR661 pKa = 11.84SISVDD666 pKa = 3.28LEE668 pKa = 4.24SVRR671 pKa = 11.84SHH673 pKa = 6.78SVDD676 pKa = 3.18GDD678 pKa = 3.74DD679 pKa = 6.07DD680 pKa = 3.87KK681 pKa = 11.81TPTQSQNLRR690 pKa = 11.84KK691 pKa = 9.91RR692 pKa = 11.84FDD694 pKa = 3.59FSVLQKK700 pKa = 11.04AVDD703 pKa = 3.84EE704 pKa = 4.55KK705 pKa = 11.42KK706 pKa = 9.88MPGSYY711 pKa = 10.46EE712 pKa = 3.95STPEE716 pKa = 3.74KK717 pKa = 10.3TEE719 pKa = 3.69PTVTVEE725 pKa = 4.69KK726 pKa = 10.64IPGVKK731 pKa = 9.67SSMGVSEE738 pKa = 4.42EE739 pKa = 4.11VEE741 pKa = 3.72NDD743 pKa = 2.43KK744 pKa = 11.08RR745 pKa = 11.84AIYY748 pKa = 9.92KK749 pKa = 10.3ADD751 pKa = 3.72IIGSDD756 pKa = 4.14RR757 pKa = 11.84INGISAVNFHH767 pKa = 6.54KK768 pKa = 10.73LFRR771 pKa = 11.84EE772 pKa = 4.03RR773 pKa = 11.84FDD775 pKa = 3.75EE776 pKa = 4.39KK777 pKa = 10.74QVSPSVMARR786 pKa = 11.84LMAVLKK792 pKa = 10.76NVGVRR797 pKa = 11.84VNIAAMTADD806 pKa = 5.05EE807 pKa = 4.85INALMKK813 pKa = 10.24MRR815 pKa = 11.84DD816 pKa = 3.74GYY818 pKa = 11.1DD819 pKa = 2.77RR820 pKa = 11.84SYY822 pKa = 11.0RR823 pKa = 11.84SSEE826 pKa = 3.89SGHH829 pKa = 5.04NRR831 pKa = 11.84EE832 pKa = 3.98FRR834 pKa = 11.84EE835 pKa = 3.57PDD837 pKa = 3.13GRR839 pKa = 11.84VLTMEE844 pKa = 3.87INLRR848 pKa = 11.84RR849 pKa = 11.84MRR851 pKa = 11.84CDD853 pKa = 3.3RR854 pKa = 11.84DD855 pKa = 3.48KK856 pKa = 11.54KK857 pKa = 10.09PLRR860 pKa = 11.84EE861 pKa = 4.02ADD863 pKa = 3.67EE864 pKa = 4.54TGLIPDD870 pKa = 3.78HH871 pKa = 6.2MARR874 pKa = 11.84KK875 pKa = 9.77LGRR878 pKa = 11.84EE879 pKa = 4.0FFMAHH884 pKa = 6.66HH885 pKa = 6.47EE886 pKa = 4.01RR887 pKa = 11.84DD888 pKa = 3.55MILARR893 pKa = 11.84VVV895 pKa = 2.92
Molecular weight: 100.35 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3533 |
713 |
1086 |
883.3 |
99.85 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.935 ± 0.319 | 1.755 ± 0.26 |
6.595 ± 0.138 | 7.614 ± 0.422 |
3.085 ± 0.382 | 6.453 ± 0.201 |
2.123 ± 0.193 | 4.812 ± 0.129 |
6.482 ± 0.224 | 7.444 ± 0.372 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
5.095 ± 0.218 | 4.217 ± 0.214 |
3.029 ± 0.275 | 1.811 ± 0.217 |
6.736 ± 0.376 | 7.416 ± 0.944 |
5.067 ± 0.39 | 8.491 ± 0.128 |
1.642 ± 0.316 | 3.198 ± 0.238 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
