
Sinorhizobium sp. RAC02
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Rhizobiaceae; Sinorhizobium/Ensifer group; Sinorhizobium; unclassified Sinorhizobium
Average proteome isoelectric point is 6.41
Get precalculated fractions of proteins
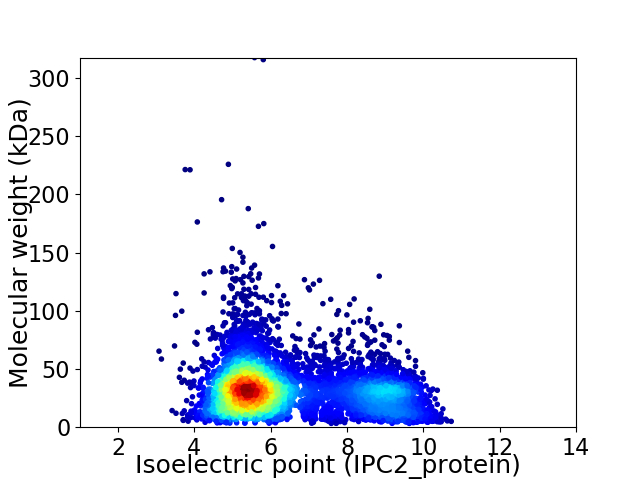
Virtual 2D-PAGE plot for 6245 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1B3M821|A0A1B3M821_9RHIZ DoxX family protein OS=Sinorhizobium sp. RAC02 OX=1842534 GN=BSY16_1827 PE=4 SV=1
MM1 pKa = 7.72PGYY4 pKa = 9.27STVEE8 pKa = 4.03GNDD11 pKa = 3.41FANVIVGQSWVVTNWQTGAGYY32 pKa = 10.96YY33 pKa = 10.29DD34 pKa = 3.6DD35 pKa = 5.91NYY37 pKa = 10.74WFDD40 pKa = 3.66TIYY43 pKa = 10.78GYY45 pKa = 11.13GGNDD49 pKa = 3.1KK50 pKa = 11.08LYY52 pKa = 11.1GLGGNDD58 pKa = 3.17RR59 pKa = 11.84LYY61 pKa = 11.28GGDD64 pKa = 4.63GVDD67 pKa = 3.47EE68 pKa = 4.83LYY70 pKa = 10.9GGKK73 pKa = 9.7GHH75 pKa = 7.25DD76 pKa = 4.02QLNGGAGNDD85 pKa = 3.5ILNGDD90 pKa = 4.27EE91 pKa = 5.08GDD93 pKa = 5.27DD94 pKa = 3.59ILSGGGGYY102 pKa = 10.08DD103 pKa = 3.25QLFGGAGWDD112 pKa = 3.87TFWGVDD118 pKa = 3.43GHH120 pKa = 6.74SSVMWGGAGADD131 pKa = 3.55KK132 pKa = 10.65MFGGSGSEE140 pKa = 3.9EE141 pKa = 4.37MYY143 pKa = 11.09GGDD146 pKa = 4.76DD147 pKa = 4.09ADD149 pKa = 3.75TMRR152 pKa = 11.84GSFGIDD158 pKa = 2.8TMSGDD163 pKa = 3.69NGHH166 pKa = 7.51DD167 pKa = 3.72DD168 pKa = 3.73MQGGGDD174 pKa = 4.24NDD176 pKa = 4.05TMSGGLGNDD185 pKa = 5.18LIAGDD190 pKa = 4.7AGADD194 pKa = 3.5TIFGNEE200 pKa = 3.62GRR202 pKa = 11.84DD203 pKa = 3.68NLSGGSGNDD212 pKa = 3.91TIWGDD217 pKa = 3.34GAYY220 pKa = 10.38FIAEE224 pKa = 3.93SNYY227 pKa = 8.62YY228 pKa = 10.36HH229 pKa = 7.19NDD231 pKa = 3.25IIDD234 pKa = 3.84GGTGNDD240 pKa = 3.74IIYY243 pKa = 10.59GGIGLDD249 pKa = 3.79TIKK252 pKa = 10.93GGTGRR257 pKa = 11.84DD258 pKa = 3.26TVYY261 pKa = 10.97GGTGDD266 pKa = 4.97DD267 pKa = 4.96VIDD270 pKa = 4.6SSDD273 pKa = 3.45GDD275 pKa = 3.75GGLVLILGADD285 pKa = 3.4DD286 pKa = 3.95SFYY289 pKa = 11.56GEE291 pKa = 5.25DD292 pKa = 4.27GNDD295 pKa = 3.42TIYY298 pKa = 11.13GRR300 pKa = 11.84NGNDD304 pKa = 3.13FLYY307 pKa = 10.8GGNGNDD313 pKa = 3.93YY314 pKa = 10.97LSGGQGSDD322 pKa = 3.39RR323 pKa = 11.84LSGDD327 pKa = 3.08AGFDD331 pKa = 3.4TIYY334 pKa = 10.92AGTGNDD340 pKa = 3.4TMDD343 pKa = 4.17GGSEE347 pKa = 3.87NDD349 pKa = 3.23NLYY352 pKa = 11.07GEE354 pKa = 5.05EE355 pKa = 4.29GADD358 pKa = 3.89SIRR361 pKa = 11.84GGTGNDD367 pKa = 3.31NVNGGAGNDD376 pKa = 4.19ALWGDD381 pKa = 3.85AGNDD385 pKa = 3.47GVYY388 pKa = 10.55GKK390 pKa = 10.7EE391 pKa = 4.3GDD393 pKa = 3.77DD394 pKa = 4.43QIWGGDD400 pKa = 3.69GNDD403 pKa = 5.13DD404 pKa = 3.2MWGDD408 pKa = 3.71AGNDD412 pKa = 3.17ILRR415 pKa = 11.84GGIGDD420 pKa = 4.61DD421 pKa = 3.52VLHH424 pKa = 6.85GNDD427 pKa = 4.12GDD429 pKa = 5.18DD430 pKa = 4.98ILFGDD435 pKa = 3.75QGTNTIYY442 pKa = 11.13GEE444 pKa = 4.1NGNDD448 pKa = 3.05RR449 pKa = 11.84FIASDD454 pKa = 5.15AIDD457 pKa = 4.05HH458 pKa = 6.3MNGGADD464 pKa = 3.39VDD466 pKa = 4.18TLSFSAYY473 pKa = 10.01LSVAVSVDD481 pKa = 3.61LAAGKK486 pKa = 10.29GLAGQALGDD495 pKa = 3.74TYY497 pKa = 11.62VGIEE501 pKa = 4.13NVVGGWGGDD510 pKa = 3.19RR511 pKa = 11.84LVGSAVANKK520 pKa = 10.4LLGEE524 pKa = 4.65KK525 pKa = 10.91GNDD528 pKa = 3.58TIFGGGGNDD537 pKa = 3.16RR538 pKa = 11.84LEE540 pKa = 4.95GGDD543 pKa = 3.38GHH545 pKa = 7.97DD546 pKa = 5.34GIRR549 pKa = 11.84GDD551 pKa = 4.16AGNDD555 pKa = 3.38TLYY558 pKa = 11.33GGAGNDD564 pKa = 3.49ALRR567 pKa = 11.84GGVGQDD573 pKa = 2.67IVFGGSGADD582 pKa = 3.09VFFYY586 pKa = 11.25VSLDD590 pKa = 3.47EE591 pKa = 4.7SGITAATRR599 pKa = 11.84DD600 pKa = 3.32IVRR603 pKa = 11.84DD604 pKa = 4.32FSRR607 pKa = 11.84AEE609 pKa = 3.76GDD611 pKa = 3.73RR612 pKa = 11.84INLSGIDD619 pKa = 3.59ASEE622 pKa = 4.03TANGNQAFTFRR633 pKa = 11.84GTNAFTGAAGEE644 pKa = 4.12LRR646 pKa = 11.84YY647 pKa = 7.71TASGGDD653 pKa = 3.67VYY655 pKa = 11.71VSGDD659 pKa = 3.29TDD661 pKa = 3.45GDD663 pKa = 3.63KK664 pKa = 11.27AADD667 pKa = 3.86FSILFDD673 pKa = 4.98NITSLAASDD682 pKa = 4.12FLLL685 pKa = 5.0
MM1 pKa = 7.72PGYY4 pKa = 9.27STVEE8 pKa = 4.03GNDD11 pKa = 3.41FANVIVGQSWVVTNWQTGAGYY32 pKa = 10.96YY33 pKa = 10.29DD34 pKa = 3.6DD35 pKa = 5.91NYY37 pKa = 10.74WFDD40 pKa = 3.66TIYY43 pKa = 10.78GYY45 pKa = 11.13GGNDD49 pKa = 3.1KK50 pKa = 11.08LYY52 pKa = 11.1GLGGNDD58 pKa = 3.17RR59 pKa = 11.84LYY61 pKa = 11.28GGDD64 pKa = 4.63GVDD67 pKa = 3.47EE68 pKa = 4.83LYY70 pKa = 10.9GGKK73 pKa = 9.7GHH75 pKa = 7.25DD76 pKa = 4.02QLNGGAGNDD85 pKa = 3.5ILNGDD90 pKa = 4.27EE91 pKa = 5.08GDD93 pKa = 5.27DD94 pKa = 3.59ILSGGGGYY102 pKa = 10.08DD103 pKa = 3.25QLFGGAGWDD112 pKa = 3.87TFWGVDD118 pKa = 3.43GHH120 pKa = 6.74SSVMWGGAGADD131 pKa = 3.55KK132 pKa = 10.65MFGGSGSEE140 pKa = 3.9EE141 pKa = 4.37MYY143 pKa = 11.09GGDD146 pKa = 4.76DD147 pKa = 4.09ADD149 pKa = 3.75TMRR152 pKa = 11.84GSFGIDD158 pKa = 2.8TMSGDD163 pKa = 3.69NGHH166 pKa = 7.51DD167 pKa = 3.72DD168 pKa = 3.73MQGGGDD174 pKa = 4.24NDD176 pKa = 4.05TMSGGLGNDD185 pKa = 5.18LIAGDD190 pKa = 4.7AGADD194 pKa = 3.5TIFGNEE200 pKa = 3.62GRR202 pKa = 11.84DD203 pKa = 3.68NLSGGSGNDD212 pKa = 3.91TIWGDD217 pKa = 3.34GAYY220 pKa = 10.38FIAEE224 pKa = 3.93SNYY227 pKa = 8.62YY228 pKa = 10.36HH229 pKa = 7.19NDD231 pKa = 3.25IIDD234 pKa = 3.84GGTGNDD240 pKa = 3.74IIYY243 pKa = 10.59GGIGLDD249 pKa = 3.79TIKK252 pKa = 10.93GGTGRR257 pKa = 11.84DD258 pKa = 3.26TVYY261 pKa = 10.97GGTGDD266 pKa = 4.97DD267 pKa = 4.96VIDD270 pKa = 4.6SSDD273 pKa = 3.45GDD275 pKa = 3.75GGLVLILGADD285 pKa = 3.4DD286 pKa = 3.95SFYY289 pKa = 11.56GEE291 pKa = 5.25DD292 pKa = 4.27GNDD295 pKa = 3.42TIYY298 pKa = 11.13GRR300 pKa = 11.84NGNDD304 pKa = 3.13FLYY307 pKa = 10.8GGNGNDD313 pKa = 3.93YY314 pKa = 10.97LSGGQGSDD322 pKa = 3.39RR323 pKa = 11.84LSGDD327 pKa = 3.08AGFDD331 pKa = 3.4TIYY334 pKa = 10.92AGTGNDD340 pKa = 3.4TMDD343 pKa = 4.17GGSEE347 pKa = 3.87NDD349 pKa = 3.23NLYY352 pKa = 11.07GEE354 pKa = 5.05EE355 pKa = 4.29GADD358 pKa = 3.89SIRR361 pKa = 11.84GGTGNDD367 pKa = 3.31NVNGGAGNDD376 pKa = 4.19ALWGDD381 pKa = 3.85AGNDD385 pKa = 3.47GVYY388 pKa = 10.55GKK390 pKa = 10.7EE391 pKa = 4.3GDD393 pKa = 3.77DD394 pKa = 4.43QIWGGDD400 pKa = 3.69GNDD403 pKa = 5.13DD404 pKa = 3.2MWGDD408 pKa = 3.71AGNDD412 pKa = 3.17ILRR415 pKa = 11.84GGIGDD420 pKa = 4.61DD421 pKa = 3.52VLHH424 pKa = 6.85GNDD427 pKa = 4.12GDD429 pKa = 5.18DD430 pKa = 4.98ILFGDD435 pKa = 3.75QGTNTIYY442 pKa = 11.13GEE444 pKa = 4.1NGNDD448 pKa = 3.05RR449 pKa = 11.84FIASDD454 pKa = 5.15AIDD457 pKa = 4.05HH458 pKa = 6.3MNGGADD464 pKa = 3.39VDD466 pKa = 4.18TLSFSAYY473 pKa = 10.01LSVAVSVDD481 pKa = 3.61LAAGKK486 pKa = 10.29GLAGQALGDD495 pKa = 3.74TYY497 pKa = 11.62VGIEE501 pKa = 4.13NVVGGWGGDD510 pKa = 3.19RR511 pKa = 11.84LVGSAVANKK520 pKa = 10.4LLGEE524 pKa = 4.65KK525 pKa = 10.91GNDD528 pKa = 3.58TIFGGGGNDD537 pKa = 3.16RR538 pKa = 11.84LEE540 pKa = 4.95GGDD543 pKa = 3.38GHH545 pKa = 7.97DD546 pKa = 5.34GIRR549 pKa = 11.84GDD551 pKa = 4.16AGNDD555 pKa = 3.38TLYY558 pKa = 11.33GGAGNDD564 pKa = 3.49ALRR567 pKa = 11.84GGVGQDD573 pKa = 2.67IVFGGSGADD582 pKa = 3.09VFFYY586 pKa = 11.25VSLDD590 pKa = 3.47EE591 pKa = 4.7SGITAATRR599 pKa = 11.84DD600 pKa = 3.32IVRR603 pKa = 11.84DD604 pKa = 4.32FSRR607 pKa = 11.84AEE609 pKa = 3.76GDD611 pKa = 3.73RR612 pKa = 11.84INLSGIDD619 pKa = 3.59ASEE622 pKa = 4.03TANGNQAFTFRR633 pKa = 11.84GTNAFTGAAGEE644 pKa = 4.12LRR646 pKa = 11.84YY647 pKa = 7.71TASGGDD653 pKa = 3.67VYY655 pKa = 11.71VSGDD659 pKa = 3.29TDD661 pKa = 3.45GDD663 pKa = 3.63KK664 pKa = 11.27AADD667 pKa = 3.86FSILFDD673 pKa = 4.98NITSLAASDD682 pKa = 4.12FLLL685 pKa = 5.0
Molecular weight: 69.79 kDa
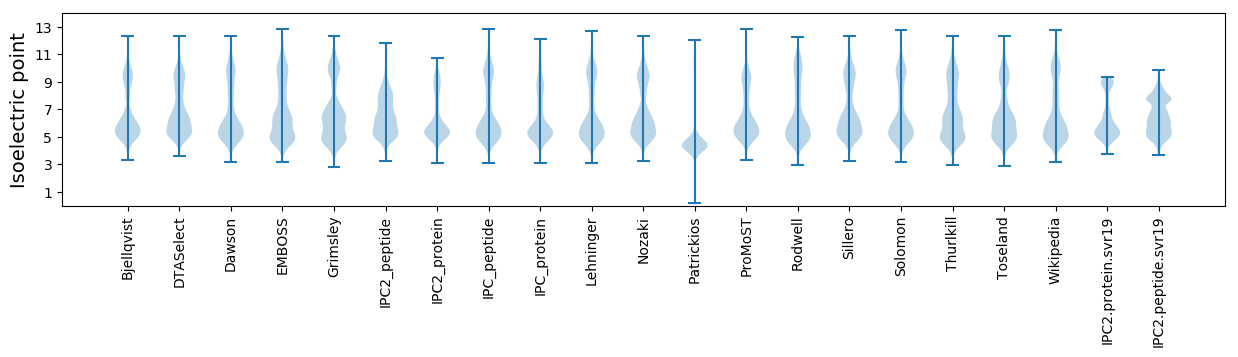
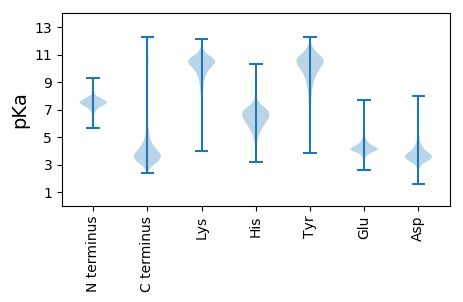
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1B3MBC2|A0A1B3MBC2_9RHIZ Methylenetetrahydrofolate--tRNA-(uracil-5-)-methyltransferase TrmFO OS=Sinorhizobium sp. RAC02 OX=1842534 GN=trmFO PE=3 SV=1
MM1 pKa = 7.82RR2 pKa = 11.84KK3 pKa = 8.97FVLVSIALATAIGSFATPSLAQKK26 pKa = 10.19SGRR29 pKa = 11.84GDD31 pKa = 3.46NGNDD35 pKa = 3.12RR36 pKa = 11.84SDD38 pKa = 3.47RR39 pKa = 11.84GSSGRR44 pKa = 11.84GEE46 pKa = 4.27YY47 pKa = 10.4YY48 pKa = 9.02RR49 pKa = 11.84TVEE52 pKa = 4.25RR53 pKa = 11.84EE54 pKa = 3.83PFTHH58 pKa = 5.37VRR60 pKa = 11.84KK61 pKa = 9.13KK62 pKa = 10.42HH63 pKa = 5.2RR64 pKa = 11.84VILANSGAYY73 pKa = 10.18CSTNWAVLFDD83 pKa = 4.24RR84 pKa = 11.84SGHH87 pKa = 4.31RR88 pKa = 11.84TKK90 pKa = 10.48VRR92 pKa = 11.84HH93 pKa = 6.07CDD95 pKa = 3.09DD96 pKa = 4.35RR97 pKa = 11.84LPIDD101 pKa = 3.45VRR103 pKa = 3.81
MM1 pKa = 7.82RR2 pKa = 11.84KK3 pKa = 8.97FVLVSIALATAIGSFATPSLAQKK26 pKa = 10.19SGRR29 pKa = 11.84GDD31 pKa = 3.46NGNDD35 pKa = 3.12RR36 pKa = 11.84SDD38 pKa = 3.47RR39 pKa = 11.84GSSGRR44 pKa = 11.84GEE46 pKa = 4.27YY47 pKa = 10.4YY48 pKa = 9.02RR49 pKa = 11.84TVEE52 pKa = 4.25RR53 pKa = 11.84EE54 pKa = 3.83PFTHH58 pKa = 5.37VRR60 pKa = 11.84KK61 pKa = 9.13KK62 pKa = 10.42HH63 pKa = 5.2RR64 pKa = 11.84VILANSGAYY73 pKa = 10.18CSTNWAVLFDD83 pKa = 4.24RR84 pKa = 11.84SGHH87 pKa = 4.31RR88 pKa = 11.84TKK90 pKa = 10.48VRR92 pKa = 11.84HH93 pKa = 6.07CDD95 pKa = 3.09DD96 pKa = 4.35RR97 pKa = 11.84LPIDD101 pKa = 3.45VRR103 pKa = 3.81
Molecular weight: 11.55 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1946794 |
29 |
2876 |
311.7 |
33.83 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.224 ± 0.04 | 0.79 ± 0.009 |
5.725 ± 0.027 | 5.749 ± 0.029 |
3.95 ± 0.023 | 8.552 ± 0.031 |
2.003 ± 0.014 | 5.571 ± 0.025 |
3.597 ± 0.025 | 10.035 ± 0.038 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.526 ± 0.016 | 2.767 ± 0.02 |
4.805 ± 0.02 | 2.926 ± 0.018 |
6.653 ± 0.031 | 5.533 ± 0.023 |
5.541 ± 0.022 | 7.477 ± 0.026 |
1.28 ± 0.013 | 2.295 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
