
Pseudidiomarina taiwanensis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Alteromonadales; Idiomarinaceae; Pseudidiomarina
Average proteome isoelectric point is 6.06
Get precalculated fractions of proteins
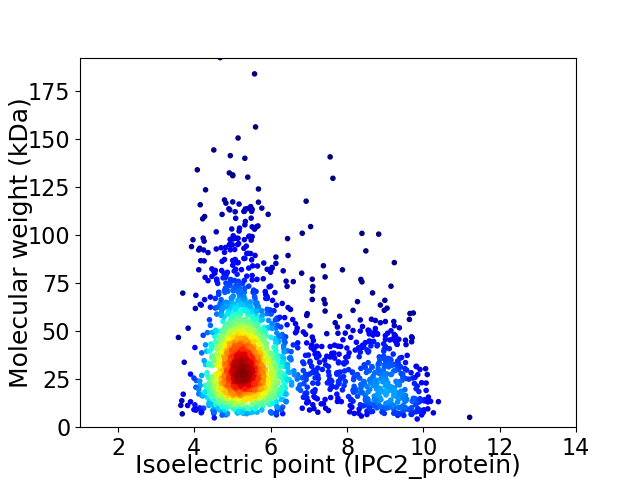
Virtual 2D-PAGE plot for 2021 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A432ZL29|A0A432ZL29_9GAMM Rhomboid family intramembrane serine protease OS=Pseudidiomarina taiwanensis OX=337250 GN=CWI83_06420 PE=4 SV=1
MM1 pKa = 7.37KK2 pKa = 10.78GIMKK6 pKa = 10.48YY7 pKa = 10.86GLATVAAVLLSACGSSSDD25 pKa = 4.55DD26 pKa = 3.32EE27 pKa = 4.77STVVLPEE34 pKa = 5.26LITEE38 pKa = 4.28VRR40 pKa = 11.84VHH42 pKa = 5.8HH43 pKa = 6.15TVADD47 pKa = 3.94APNVNVLVNGGAAVEE62 pKa = 4.14DD63 pKa = 4.0AGFGASSTVLEE74 pKa = 4.89LDD76 pKa = 3.13AGTYY80 pKa = 9.93SLQVDD85 pKa = 5.08AILADD90 pKa = 3.92GSTATVIGPVNDD102 pKa = 3.43TLAANTRR109 pKa = 11.84YY110 pKa = 10.09EE111 pKa = 3.68ILAIGKK117 pKa = 9.01VADD120 pKa = 3.86EE121 pKa = 4.79TIQPLIITNLNSEE134 pKa = 4.22IADD137 pKa = 3.53GNARR141 pKa = 11.84LQVVHH146 pKa = 7.12AAPDD150 pKa = 3.47APLVDD155 pKa = 4.55IYY157 pKa = 10.72LTAPDD162 pKa = 4.75ADD164 pKa = 4.44LSASSPAATLAFSDD178 pKa = 3.56FTGQVEE184 pKa = 4.82VPAGDD189 pKa = 3.56YY190 pKa = 9.08QVRR193 pKa = 11.84ITPAGDD199 pKa = 3.2PATVVFDD206 pKa = 4.33SGTLPLTAGADD217 pKa = 4.09LIVSAVTNTKK227 pKa = 10.68AGDD230 pKa = 4.09SPVMLQVADD239 pKa = 4.09GSGASIVTDD248 pKa = 3.73INAGADD254 pKa = 3.36VRR256 pKa = 11.84IVHH259 pKa = 6.51SSADD263 pKa = 3.55APTVDD268 pKa = 5.59FVADD272 pKa = 3.3GDD274 pKa = 4.24MPATLATLSFGQFTDD289 pKa = 3.71YY290 pKa = 11.54LNVADD295 pKa = 5.45GDD297 pKa = 4.1YY298 pKa = 11.1LIDD301 pKa = 4.11VIVNGTTTVALNDD314 pKa = 3.64IPLSLAQGEE323 pKa = 4.63IYY325 pKa = 10.33SVYY328 pKa = 10.69AIGSAGDD335 pKa = 3.68GSLTAGVLTPDD346 pKa = 3.26TRR348 pKa = 11.84RR349 pKa = 11.84IATEE353 pKa = 3.67AKK355 pKa = 9.37LQAVHH360 pKa = 7.15ASPTAGDD367 pKa = 3.11VDD369 pKa = 3.89IYY371 pKa = 10.55VTATDD376 pKa = 5.67DD377 pKa = 3.47ITSATPAFTAVPFTTEE393 pKa = 4.08NGPASTGYY401 pKa = 10.26VSLMPGDD408 pKa = 4.28YY409 pKa = 10.49YY410 pKa = 10.86VTITPTGTKK419 pKa = 9.83DD420 pKa = 3.13AAIGPEE426 pKa = 4.21LVTLEE431 pKa = 4.14GNGIYY436 pKa = 9.51TAVAVDD442 pKa = 3.61AVGGGVPLNVIAMDD456 pKa = 4.59DD457 pKa = 3.9FATVDD462 pKa = 3.14
MM1 pKa = 7.37KK2 pKa = 10.78GIMKK6 pKa = 10.48YY7 pKa = 10.86GLATVAAVLLSACGSSSDD25 pKa = 4.55DD26 pKa = 3.32EE27 pKa = 4.77STVVLPEE34 pKa = 5.26LITEE38 pKa = 4.28VRR40 pKa = 11.84VHH42 pKa = 5.8HH43 pKa = 6.15TVADD47 pKa = 3.94APNVNVLVNGGAAVEE62 pKa = 4.14DD63 pKa = 4.0AGFGASSTVLEE74 pKa = 4.89LDD76 pKa = 3.13AGTYY80 pKa = 9.93SLQVDD85 pKa = 5.08AILADD90 pKa = 3.92GSTATVIGPVNDD102 pKa = 3.43TLAANTRR109 pKa = 11.84YY110 pKa = 10.09EE111 pKa = 3.68ILAIGKK117 pKa = 9.01VADD120 pKa = 3.86EE121 pKa = 4.79TIQPLIITNLNSEE134 pKa = 4.22IADD137 pKa = 3.53GNARR141 pKa = 11.84LQVVHH146 pKa = 7.12AAPDD150 pKa = 3.47APLVDD155 pKa = 4.55IYY157 pKa = 10.72LTAPDD162 pKa = 4.75ADD164 pKa = 4.44LSASSPAATLAFSDD178 pKa = 3.56FTGQVEE184 pKa = 4.82VPAGDD189 pKa = 3.56YY190 pKa = 9.08QVRR193 pKa = 11.84ITPAGDD199 pKa = 3.2PATVVFDD206 pKa = 4.33SGTLPLTAGADD217 pKa = 4.09LIVSAVTNTKK227 pKa = 10.68AGDD230 pKa = 4.09SPVMLQVADD239 pKa = 4.09GSGASIVTDD248 pKa = 3.73INAGADD254 pKa = 3.36VRR256 pKa = 11.84IVHH259 pKa = 6.51SSADD263 pKa = 3.55APTVDD268 pKa = 5.59FVADD272 pKa = 3.3GDD274 pKa = 4.24MPATLATLSFGQFTDD289 pKa = 3.71YY290 pKa = 11.54LNVADD295 pKa = 5.45GDD297 pKa = 4.1YY298 pKa = 11.1LIDD301 pKa = 4.11VIVNGTTTVALNDD314 pKa = 3.64IPLSLAQGEE323 pKa = 4.63IYY325 pKa = 10.33SVYY328 pKa = 10.69AIGSAGDD335 pKa = 3.68GSLTAGVLTPDD346 pKa = 3.26TRR348 pKa = 11.84RR349 pKa = 11.84IATEE353 pKa = 3.67AKK355 pKa = 9.37LQAVHH360 pKa = 7.15ASPTAGDD367 pKa = 3.11VDD369 pKa = 3.89IYY371 pKa = 10.55VTATDD376 pKa = 5.67DD377 pKa = 3.47ITSATPAFTAVPFTTEE393 pKa = 4.08NGPASTGYY401 pKa = 10.26VSLMPGDD408 pKa = 4.28YY409 pKa = 10.49YY410 pKa = 10.86VTITPTGTKK419 pKa = 9.83DD420 pKa = 3.13AAIGPEE426 pKa = 4.21LVTLEE431 pKa = 4.14GNGIYY436 pKa = 9.51TAVAVDD442 pKa = 3.61AVGGGVPLNVIAMDD456 pKa = 4.59DD457 pKa = 3.9FATVDD462 pKa = 3.14
Molecular weight: 46.74 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A432ZEQ9|A0A432ZEQ9_9GAMM DUF2489 domain-containing protein OS=Pseudidiomarina taiwanensis OX=337250 GN=CWI83_08455 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.49RR12 pKa = 11.84KK13 pKa = 7.97RR14 pKa = 11.84THH16 pKa = 5.89GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.4NGRR28 pKa = 11.84QVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.94GRR39 pKa = 11.84KK40 pKa = 8.67VLSAA44 pKa = 4.05
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.49RR12 pKa = 11.84KK13 pKa = 7.97RR14 pKa = 11.84THH16 pKa = 5.89GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.4NGRR28 pKa = 11.84QVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.94GRR39 pKa = 11.84KK40 pKa = 8.67VLSAA44 pKa = 4.05
Molecular weight: 5.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
682925 |
37 |
1759 |
337.9 |
37.46 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.358 ± 0.061 | 0.858 ± 0.016 |
5.393 ± 0.046 | 6.354 ± 0.051 |
3.877 ± 0.035 | 6.978 ± 0.051 |
2.273 ± 0.027 | 5.665 ± 0.041 |
3.904 ± 0.041 | 10.837 ± 0.069 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.365 ± 0.027 | 3.369 ± 0.032 |
4.292 ± 0.034 | 5.578 ± 0.06 |
5.669 ± 0.041 | 5.637 ± 0.036 |
5.121 ± 0.03 | 7.144 ± 0.045 |
1.367 ± 0.028 | 2.962 ± 0.033 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
