
Croceitalea dokdonensis DOKDO 023
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Croceitalea; Croceitalea dokdonensis
Average proteome isoelectric point is 6.78
Get precalculated fractions of proteins
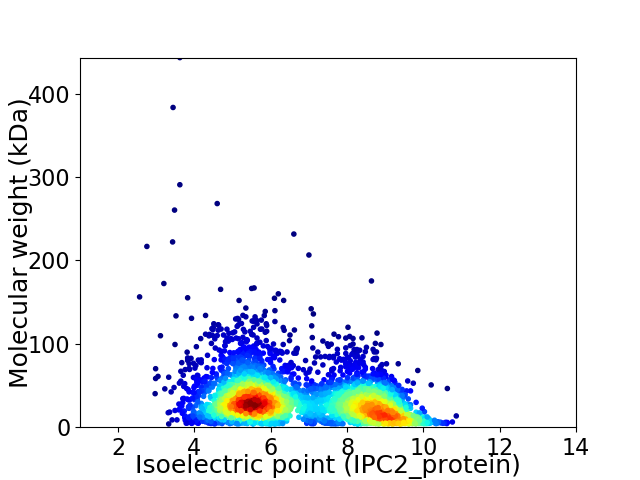
Virtual 2D-PAGE plot for 3682 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0P7AD04|A0A0P7AD04_9FLAO 3'-5' exonuclease OS=Croceitalea dokdonensis DOKDO 023 OX=1300341 GN=I595_3051 PE=4 SV=1
MM1 pKa = 7.74LMPMVLRR8 pKa = 11.84FFAVISVFLLLSCGGDD24 pKa = 3.7DD25 pKa = 4.12EE26 pKa = 5.72PEE28 pKa = 4.05NQMTTSVTVAFLAQEE43 pKa = 4.44TTGSEE48 pKa = 3.88ALGNNLPSVSVRR60 pKa = 11.84GTVASEE66 pKa = 4.07VSVSVLLTNTGTATLGEE83 pKa = 4.69DD84 pKa = 3.98FSFASPTVISIPIGNYY100 pKa = 10.23DD101 pKa = 3.6GTALTAIPLPGLSILDD117 pKa = 3.66DD118 pKa = 3.78TVQEE122 pKa = 4.21GFEE125 pKa = 4.3TIIFSLDD132 pKa = 3.26NPTEE136 pKa = 4.2GVNLGDD142 pKa = 3.91VATTTYY148 pKa = 10.02TINDD152 pKa = 4.14DD153 pKa = 3.46EE154 pKa = 4.59TVQVGFSQAMASDD167 pKa = 4.1RR168 pKa = 11.84EE169 pKa = 4.39NVGGNLPTLFVTGTLSEE186 pKa = 4.88DD187 pKa = 3.11KK188 pKa = 10.49TITLTAVDD196 pKa = 4.59SGDD199 pKa = 3.3ATLGQDD205 pKa = 3.7YY206 pKa = 10.73NVASPLVVTIPAGTYY221 pKa = 10.54DD222 pKa = 3.78GTTATALTIPALAIIDD238 pKa = 3.75DD239 pKa = 4.3TVPEE243 pKa = 4.04EE244 pKa = 4.38SEE246 pKa = 4.48SFEE249 pKa = 4.41LTLSDD254 pKa = 4.3PSNGLLLNEE263 pKa = 5.53LDD265 pKa = 3.11RR266 pKa = 11.84TMYY269 pKa = 9.87TIEE272 pKa = 4.28GVAPCEE278 pKa = 3.83NGRR281 pKa = 11.84ANGFPCNGFDD291 pKa = 3.51LLARR295 pKa = 11.84MEE297 pKa = 4.56HH298 pKa = 5.88STFSSTQGNDD308 pKa = 2.03IWGWTDD314 pKa = 2.76TATGHH319 pKa = 6.31EE320 pKa = 4.43YY321 pKa = 11.41ALVALNNGTAFVDD334 pKa = 3.75ITADD338 pKa = 3.53DD339 pKa = 3.88PVYY342 pKa = 10.65LGKK345 pKa = 10.66LPSATGSSVWRR356 pKa = 11.84DD357 pKa = 3.0VKK359 pKa = 10.8IYY361 pKa = 10.63GNHH364 pKa = 6.38AYY366 pKa = 9.27IVSEE370 pKa = 4.01ASGHH374 pKa = 4.89GMQVFDD380 pKa = 4.03LTRR383 pKa = 11.84LRR385 pKa = 11.84NVANPPTDD393 pKa = 3.66FTQDD397 pKa = 2.62ARR399 pKa = 11.84YY400 pKa = 7.96TDD402 pKa = 3.22IGNAHH407 pKa = 5.85NVVINEE413 pKa = 4.15QNGFAYY419 pKa = 9.56PVGTARR425 pKa = 11.84NDD427 pKa = 3.55TFNGGVHH434 pKa = 6.47FVDD437 pKa = 3.65IQNPTSPTAAGGYY450 pKa = 9.4GDD452 pKa = 4.67NGYY455 pKa = 7.73THH457 pKa = 7.89DD458 pKa = 4.1AQVVNYY464 pKa = 7.53EE465 pKa = 4.58GPDD468 pKa = 3.02ADD470 pKa = 3.79YY471 pKa = 10.26TGRR474 pKa = 11.84EE475 pKa = 3.78IFIGANEE482 pKa = 4.01DD483 pKa = 3.19QIAIVDD489 pKa = 3.81ITDD492 pKa = 3.47KK493 pKa = 11.48ANPTEE498 pKa = 4.22IATITYY504 pKa = 9.87GNLGYY509 pKa = 7.89THH511 pKa = 6.47QGWFTEE517 pKa = 4.08DD518 pKa = 2.99QRR520 pKa = 11.84FFLLGDD526 pKa = 3.76EE527 pKa = 4.73LDD529 pKa = 4.53EE530 pKa = 5.86IDD532 pKa = 4.86FGFNSRR538 pKa = 11.84TLVFDD543 pKa = 5.01LQDD546 pKa = 3.54LDD548 pKa = 4.28NPVLHH553 pKa = 5.73TTYY556 pKa = 10.66LGPTAAIDD564 pKa = 3.48HH565 pKa = 6.55NGYY568 pKa = 9.22VHH570 pKa = 7.32GDD572 pKa = 3.37EE573 pKa = 5.46FFLANYY579 pKa = 6.66TAGVRR584 pKa = 11.84VLDD587 pKa = 4.27LSDD590 pKa = 3.65IEE592 pKa = 4.76GGNLTEE598 pKa = 5.23IGFFDD603 pKa = 4.21TFPDD607 pKa = 3.33NDD609 pKa = 3.37VASFNGVWSVYY620 pKa = 10.4PYY622 pKa = 10.59FEE624 pKa = 4.12SGKK627 pKa = 10.47IIVNDD632 pKa = 3.37SDD634 pKa = 3.46TGFFIIQKK642 pKa = 8.69SQQ644 pKa = 2.6
MM1 pKa = 7.74LMPMVLRR8 pKa = 11.84FFAVISVFLLLSCGGDD24 pKa = 3.7DD25 pKa = 4.12EE26 pKa = 5.72PEE28 pKa = 4.05NQMTTSVTVAFLAQEE43 pKa = 4.44TTGSEE48 pKa = 3.88ALGNNLPSVSVRR60 pKa = 11.84GTVASEE66 pKa = 4.07VSVSVLLTNTGTATLGEE83 pKa = 4.69DD84 pKa = 3.98FSFASPTVISIPIGNYY100 pKa = 10.23DD101 pKa = 3.6GTALTAIPLPGLSILDD117 pKa = 3.66DD118 pKa = 3.78TVQEE122 pKa = 4.21GFEE125 pKa = 4.3TIIFSLDD132 pKa = 3.26NPTEE136 pKa = 4.2GVNLGDD142 pKa = 3.91VATTTYY148 pKa = 10.02TINDD152 pKa = 4.14DD153 pKa = 3.46EE154 pKa = 4.59TVQVGFSQAMASDD167 pKa = 4.1RR168 pKa = 11.84EE169 pKa = 4.39NVGGNLPTLFVTGTLSEE186 pKa = 4.88DD187 pKa = 3.11KK188 pKa = 10.49TITLTAVDD196 pKa = 4.59SGDD199 pKa = 3.3ATLGQDD205 pKa = 3.7YY206 pKa = 10.73NVASPLVVTIPAGTYY221 pKa = 10.54DD222 pKa = 3.78GTTATALTIPALAIIDD238 pKa = 3.75DD239 pKa = 4.3TVPEE243 pKa = 4.04EE244 pKa = 4.38SEE246 pKa = 4.48SFEE249 pKa = 4.41LTLSDD254 pKa = 4.3PSNGLLLNEE263 pKa = 5.53LDD265 pKa = 3.11RR266 pKa = 11.84TMYY269 pKa = 9.87TIEE272 pKa = 4.28GVAPCEE278 pKa = 3.83NGRR281 pKa = 11.84ANGFPCNGFDD291 pKa = 3.51LLARR295 pKa = 11.84MEE297 pKa = 4.56HH298 pKa = 5.88STFSSTQGNDD308 pKa = 2.03IWGWTDD314 pKa = 2.76TATGHH319 pKa = 6.31EE320 pKa = 4.43YY321 pKa = 11.41ALVALNNGTAFVDD334 pKa = 3.75ITADD338 pKa = 3.53DD339 pKa = 3.88PVYY342 pKa = 10.65LGKK345 pKa = 10.66LPSATGSSVWRR356 pKa = 11.84DD357 pKa = 3.0VKK359 pKa = 10.8IYY361 pKa = 10.63GNHH364 pKa = 6.38AYY366 pKa = 9.27IVSEE370 pKa = 4.01ASGHH374 pKa = 4.89GMQVFDD380 pKa = 4.03LTRR383 pKa = 11.84LRR385 pKa = 11.84NVANPPTDD393 pKa = 3.66FTQDD397 pKa = 2.62ARR399 pKa = 11.84YY400 pKa = 7.96TDD402 pKa = 3.22IGNAHH407 pKa = 5.85NVVINEE413 pKa = 4.15QNGFAYY419 pKa = 9.56PVGTARR425 pKa = 11.84NDD427 pKa = 3.55TFNGGVHH434 pKa = 6.47FVDD437 pKa = 3.65IQNPTSPTAAGGYY450 pKa = 9.4GDD452 pKa = 4.67NGYY455 pKa = 7.73THH457 pKa = 7.89DD458 pKa = 4.1AQVVNYY464 pKa = 7.53EE465 pKa = 4.58GPDD468 pKa = 3.02ADD470 pKa = 3.79YY471 pKa = 10.26TGRR474 pKa = 11.84EE475 pKa = 3.78IFIGANEE482 pKa = 4.01DD483 pKa = 3.19QIAIVDD489 pKa = 3.81ITDD492 pKa = 3.47KK493 pKa = 11.48ANPTEE498 pKa = 4.22IATITYY504 pKa = 9.87GNLGYY509 pKa = 7.89THH511 pKa = 6.47QGWFTEE517 pKa = 4.08DD518 pKa = 2.99QRR520 pKa = 11.84FFLLGDD526 pKa = 3.76EE527 pKa = 4.73LDD529 pKa = 4.53EE530 pKa = 5.86IDD532 pKa = 4.86FGFNSRR538 pKa = 11.84TLVFDD543 pKa = 5.01LQDD546 pKa = 3.54LDD548 pKa = 4.28NPVLHH553 pKa = 5.73TTYY556 pKa = 10.66LGPTAAIDD564 pKa = 3.48HH565 pKa = 6.55NGYY568 pKa = 9.22VHH570 pKa = 7.32GDD572 pKa = 3.37EE573 pKa = 5.46FFLANYY579 pKa = 6.66TAGVRR584 pKa = 11.84VLDD587 pKa = 4.27LSDD590 pKa = 3.65IEE592 pKa = 4.76GGNLTEE598 pKa = 5.23IGFFDD603 pKa = 4.21TFPDD607 pKa = 3.33NDD609 pKa = 3.37VASFNGVWSVYY620 pKa = 10.4PYY622 pKa = 10.59FEE624 pKa = 4.12SGKK627 pKa = 10.47IIVNDD632 pKa = 3.37SDD634 pKa = 3.46TGFFIIQKK642 pKa = 8.69SQQ644 pKa = 2.6
Molecular weight: 69.09 kDa
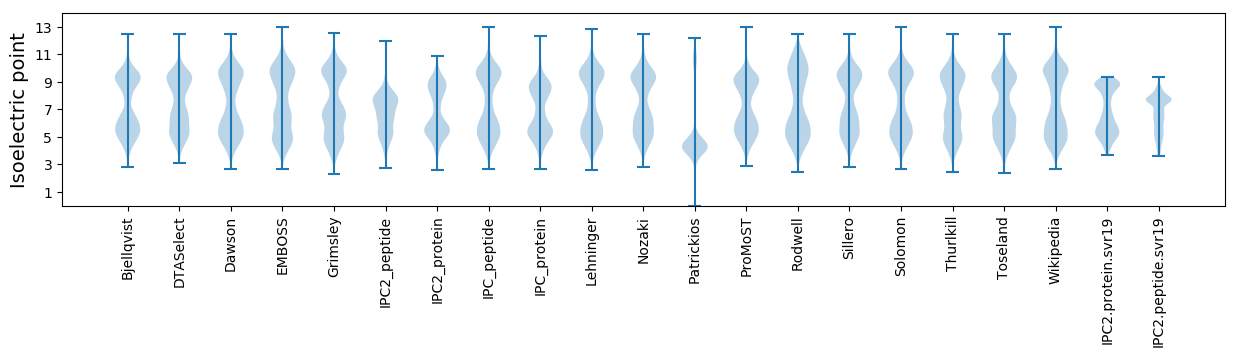
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0P7B2B5|A0A0P7B2B5_9FLAO Uncharacterized protein OS=Croceitalea dokdonensis DOKDO 023 OX=1300341 GN=I595_1994 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.59RR3 pKa = 11.84TFQPSKK9 pKa = 9.13RR10 pKa = 11.84KK11 pKa = 9.48RR12 pKa = 11.84RR13 pKa = 11.84NKK15 pKa = 9.49HH16 pKa = 3.94GFRR19 pKa = 11.84EE20 pKa = 4.27RR21 pKa = 11.84MASANGRR28 pKa = 11.84KK29 pKa = 9.04VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.08GRR39 pKa = 11.84KK40 pKa = 8.3KK41 pKa = 9.83LTVSSEE47 pKa = 3.9PRR49 pKa = 11.84HH50 pKa = 5.77KK51 pKa = 10.61KK52 pKa = 9.84
MM1 pKa = 7.45KK2 pKa = 9.59RR3 pKa = 11.84TFQPSKK9 pKa = 9.13RR10 pKa = 11.84KK11 pKa = 9.48RR12 pKa = 11.84RR13 pKa = 11.84NKK15 pKa = 9.49HH16 pKa = 3.94GFRR19 pKa = 11.84EE20 pKa = 4.27RR21 pKa = 11.84MASANGRR28 pKa = 11.84KK29 pKa = 9.04VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.08GRR39 pKa = 11.84KK40 pKa = 8.3KK41 pKa = 9.83LTVSSEE47 pKa = 3.9PRR49 pKa = 11.84HH50 pKa = 5.77KK51 pKa = 10.61KK52 pKa = 9.84
Molecular weight: 6.24 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1160232 |
37 |
4185 |
315.1 |
35.41 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.12 ± 0.039 | 0.753 ± 0.014 |
5.604 ± 0.047 | 6.296 ± 0.039 |
5.225 ± 0.038 | 6.921 ± 0.041 |
1.897 ± 0.024 | 6.952 ± 0.033 |
7.094 ± 0.069 | 9.673 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.338 ± 0.022 | 5.572 ± 0.052 |
3.686 ± 0.028 | 3.744 ± 0.023 |
3.854 ± 0.03 | 5.933 ± 0.033 |
5.898 ± 0.057 | 6.446 ± 0.031 |
1.143 ± 0.016 | 3.85 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
