
Equus caballus (Horse)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Deuterostomia; Chordata; Craniata; Vertebrata; Gnathostomata; Teleostomi; Euteleostomi; Sarcopterygii; Dipnotetrapodomorpha; Tetrapoda; Amniota; Mammalia; Theria; Eutheria; Boreoeutheria;
Average proteome isoelectric point is 6.78
Get precalculated fractions of proteins
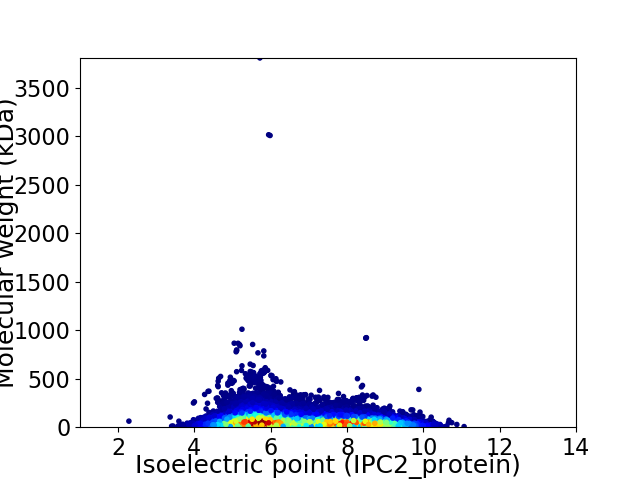
Virtual 2D-PAGE plot for 44491 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A3Q2LMH3|A0A3Q2LMH3_HORSE Prostaglandin F synthase 1-like OS=Equus caballus OX=9796 GN=LOC111771772 PE=3 SV=1
II1 pKa = 6.74GTSQNVSTKK10 pKa = 11.02DD11 pKa = 3.34PVTSPEE17 pKa = 3.83NSEE20 pKa = 3.88TRR22 pKa = 11.84EE23 pKa = 4.01NYY25 pKa = 7.56PTDD28 pKa = 3.82DD29 pKa = 3.59VEE31 pKa = 4.43TTEE34 pKa = 4.06DD35 pKa = 4.04HH36 pKa = 6.96IIEE39 pKa = 4.23ALGTNEE45 pKa = 5.68DD46 pKa = 4.49YY47 pKa = 8.18TTEE50 pKa = 3.99PLEE53 pKa = 4.05TLEE56 pKa = 5.84DD57 pKa = 3.98YY58 pKa = 7.59TTADD62 pKa = 4.33LGPTEE67 pKa = 6.0DD68 pKa = 5.59NITDD72 pKa = 3.77DD73 pKa = 4.13LGTIEE78 pKa = 4.67DD79 pKa = 4.51HH80 pKa = 7.16VIEE83 pKa = 4.86APGTTEE89 pKa = 5.69DD90 pKa = 4.06YY91 pKa = 8.14TTEE94 pKa = 3.85HH95 pKa = 7.13LEE97 pKa = 3.78TSEE100 pKa = 4.76YY101 pKa = 9.88YY102 pKa = 7.59TTTDD106 pKa = 2.35SGAYY110 pKa = 9.85DD111 pKa = 4.62DD112 pKa = 4.53YY113 pKa = 10.32TTVALGPEE121 pKa = 4.77DD122 pKa = 4.15YY123 pKa = 7.71TTADD127 pKa = 3.56LGTTKK132 pKa = 10.87DD133 pKa = 3.49SFAADD138 pKa = 3.63LGTIEE143 pKa = 6.17DD144 pKa = 5.36YY145 pKa = 10.28ITDD148 pKa = 3.94DD149 pKa = 3.34PRR151 pKa = 11.84TTEE154 pKa = 4.89DD155 pKa = 3.81YY156 pKa = 7.79TTVDD160 pKa = 3.44LGTTEE165 pKa = 5.56DD166 pKa = 4.93SFTADD171 pKa = 3.05PGTTKK176 pKa = 10.81DD177 pKa = 3.54YY178 pKa = 7.7TTADD182 pKa = 3.57LGPTEE187 pKa = 4.55DD188 pKa = 4.55HH189 pKa = 6.87VIHH192 pKa = 6.77GPGTIEE198 pKa = 5.74DD199 pKa = 4.41YY200 pKa = 8.56TTEE203 pKa = 3.93HH204 pKa = 7.05LEE206 pKa = 4.15TPEE209 pKa = 5.91DD210 pKa = 4.29YY211 pKa = 8.69DD212 pKa = 3.9TSDD215 pKa = 3.48LGTTEE220 pKa = 5.1HH221 pKa = 6.32YY222 pKa = 7.56TTVDD226 pKa = 2.85LGTTKK231 pKa = 10.45DD232 pKa = 4.07YY233 pKa = 8.54PTDD236 pKa = 3.8DD237 pKa = 3.79PGITKK242 pKa = 10.17DD243 pKa = 4.27YY244 pKa = 9.91ITVDD248 pKa = 3.14PGTTEE253 pKa = 4.74DD254 pKa = 5.42SFTADD259 pKa = 3.24LGTTEE264 pKa = 5.89DD265 pKa = 3.9YY266 pKa = 10.42TISDD270 pKa = 4.14LGIPADD276 pKa = 3.97SLIVDD281 pKa = 4.79LGTTKK286 pKa = 10.85YY287 pKa = 9.11YY288 pKa = 8.22TTDD291 pKa = 3.21DD292 pKa = 3.75PGITKK297 pKa = 9.91EE298 pKa = 4.25YY299 pKa = 9.05ITVEE303 pKa = 4.11PGTTEE308 pKa = 4.58DD309 pKa = 5.29SFTADD314 pKa = 3.24LGTTEE319 pKa = 6.1DD320 pKa = 4.33YY321 pKa = 7.67TTSDD325 pKa = 3.84LGTTTDD331 pKa = 3.31SFIVDD336 pKa = 4.19LVTTKK341 pKa = 10.76DD342 pKa = 3.6YY343 pKa = 8.03TTDD346 pKa = 3.42DD347 pKa = 3.67PGITKK352 pKa = 10.1DD353 pKa = 3.89YY354 pKa = 10.84ISVEE358 pKa = 3.95PGTTKK363 pKa = 10.97DD364 pKa = 3.52SFTADD369 pKa = 3.23LEE371 pKa = 4.53TTEE374 pKa = 4.76DD375 pKa = 3.06HH376 pKa = 6.79TIADD380 pKa = 4.05RR381 pKa = 11.84GTTKK385 pKa = 10.92DD386 pKa = 4.12SFTTDD391 pKa = 2.6PGTTVDD397 pKa = 5.57SFTADD402 pKa = 2.95LGTTEE407 pKa = 5.88DD408 pKa = 4.21YY409 pKa = 7.68TTADD413 pKa = 3.65LRR415 pKa = 11.84ITTDD419 pKa = 2.76SFTADD424 pKa = 2.98LRR426 pKa = 11.84TTKK429 pKa = 10.51GYY431 pKa = 7.08TTDD434 pKa = 3.97DD435 pKa = 3.67PEE437 pKa = 4.02ITKK440 pKa = 10.59GYY442 pKa = 8.18ITVDD446 pKa = 3.0PGTTEE451 pKa = 4.74DD452 pKa = 5.42SFTADD457 pKa = 3.21LGTTKK462 pKa = 10.93DD463 pKa = 3.25HH464 pKa = 5.83TTADD468 pKa = 3.65LRR470 pKa = 11.84TTVYY474 pKa = 10.85SCTADD479 pKa = 3.24LGTTKK484 pKa = 10.55DD485 pKa = 3.65YY486 pKa = 7.7TTADD490 pKa = 3.49PGTTEE495 pKa = 5.71DD496 pKa = 4.26YY497 pKa = 7.59TTKK500 pKa = 10.55HH501 pKa = 6.26LNTHH505 pKa = 5.61SEE507 pKa = 4.26GTDD510 pKa = 2.84SVTAVKK516 pKa = 8.77PTGLLTPIGIILISLAATTAIVALFVGLGLIVVSIWPRR554 pKa = 11.84NVKK557 pKa = 10.17EE558 pKa = 3.79IMGMKK563 pKa = 10.17DD564 pKa = 3.12SEE566 pKa = 4.45EE567 pKa = 4.22EE568 pKa = 3.91EE569 pKa = 4.86GMGSS573 pKa = 3.24
II1 pKa = 6.74GTSQNVSTKK10 pKa = 11.02DD11 pKa = 3.34PVTSPEE17 pKa = 3.83NSEE20 pKa = 3.88TRR22 pKa = 11.84EE23 pKa = 4.01NYY25 pKa = 7.56PTDD28 pKa = 3.82DD29 pKa = 3.59VEE31 pKa = 4.43TTEE34 pKa = 4.06DD35 pKa = 4.04HH36 pKa = 6.96IIEE39 pKa = 4.23ALGTNEE45 pKa = 5.68DD46 pKa = 4.49YY47 pKa = 8.18TTEE50 pKa = 3.99PLEE53 pKa = 4.05TLEE56 pKa = 5.84DD57 pKa = 3.98YY58 pKa = 7.59TTADD62 pKa = 4.33LGPTEE67 pKa = 6.0DD68 pKa = 5.59NITDD72 pKa = 3.77DD73 pKa = 4.13LGTIEE78 pKa = 4.67DD79 pKa = 4.51HH80 pKa = 7.16VIEE83 pKa = 4.86APGTTEE89 pKa = 5.69DD90 pKa = 4.06YY91 pKa = 8.14TTEE94 pKa = 3.85HH95 pKa = 7.13LEE97 pKa = 3.78TSEE100 pKa = 4.76YY101 pKa = 9.88YY102 pKa = 7.59TTTDD106 pKa = 2.35SGAYY110 pKa = 9.85DD111 pKa = 4.62DD112 pKa = 4.53YY113 pKa = 10.32TTVALGPEE121 pKa = 4.77DD122 pKa = 4.15YY123 pKa = 7.71TTADD127 pKa = 3.56LGTTKK132 pKa = 10.87DD133 pKa = 3.49SFAADD138 pKa = 3.63LGTIEE143 pKa = 6.17DD144 pKa = 5.36YY145 pKa = 10.28ITDD148 pKa = 3.94DD149 pKa = 3.34PRR151 pKa = 11.84TTEE154 pKa = 4.89DD155 pKa = 3.81YY156 pKa = 7.79TTVDD160 pKa = 3.44LGTTEE165 pKa = 5.56DD166 pKa = 4.93SFTADD171 pKa = 3.05PGTTKK176 pKa = 10.81DD177 pKa = 3.54YY178 pKa = 7.7TTADD182 pKa = 3.57LGPTEE187 pKa = 4.55DD188 pKa = 4.55HH189 pKa = 6.87VIHH192 pKa = 6.77GPGTIEE198 pKa = 5.74DD199 pKa = 4.41YY200 pKa = 8.56TTEE203 pKa = 3.93HH204 pKa = 7.05LEE206 pKa = 4.15TPEE209 pKa = 5.91DD210 pKa = 4.29YY211 pKa = 8.69DD212 pKa = 3.9TSDD215 pKa = 3.48LGTTEE220 pKa = 5.1HH221 pKa = 6.32YY222 pKa = 7.56TTVDD226 pKa = 2.85LGTTKK231 pKa = 10.45DD232 pKa = 4.07YY233 pKa = 8.54PTDD236 pKa = 3.8DD237 pKa = 3.79PGITKK242 pKa = 10.17DD243 pKa = 4.27YY244 pKa = 9.91ITVDD248 pKa = 3.14PGTTEE253 pKa = 4.74DD254 pKa = 5.42SFTADD259 pKa = 3.24LGTTEE264 pKa = 5.89DD265 pKa = 3.9YY266 pKa = 10.42TISDD270 pKa = 4.14LGIPADD276 pKa = 3.97SLIVDD281 pKa = 4.79LGTTKK286 pKa = 10.85YY287 pKa = 9.11YY288 pKa = 8.22TTDD291 pKa = 3.21DD292 pKa = 3.75PGITKK297 pKa = 9.91EE298 pKa = 4.25YY299 pKa = 9.05ITVEE303 pKa = 4.11PGTTEE308 pKa = 4.58DD309 pKa = 5.29SFTADD314 pKa = 3.24LGTTEE319 pKa = 6.1DD320 pKa = 4.33YY321 pKa = 7.67TTSDD325 pKa = 3.84LGTTTDD331 pKa = 3.31SFIVDD336 pKa = 4.19LVTTKK341 pKa = 10.76DD342 pKa = 3.6YY343 pKa = 8.03TTDD346 pKa = 3.42DD347 pKa = 3.67PGITKK352 pKa = 10.1DD353 pKa = 3.89YY354 pKa = 10.84ISVEE358 pKa = 3.95PGTTKK363 pKa = 10.97DD364 pKa = 3.52SFTADD369 pKa = 3.23LEE371 pKa = 4.53TTEE374 pKa = 4.76DD375 pKa = 3.06HH376 pKa = 6.79TIADD380 pKa = 4.05RR381 pKa = 11.84GTTKK385 pKa = 10.92DD386 pKa = 4.12SFTTDD391 pKa = 2.6PGTTVDD397 pKa = 5.57SFTADD402 pKa = 2.95LGTTEE407 pKa = 5.88DD408 pKa = 4.21YY409 pKa = 7.68TTADD413 pKa = 3.65LRR415 pKa = 11.84ITTDD419 pKa = 2.76SFTADD424 pKa = 2.98LRR426 pKa = 11.84TTKK429 pKa = 10.51GYY431 pKa = 7.08TTDD434 pKa = 3.97DD435 pKa = 3.67PEE437 pKa = 4.02ITKK440 pKa = 10.59GYY442 pKa = 8.18ITVDD446 pKa = 3.0PGTTEE451 pKa = 4.74DD452 pKa = 5.42SFTADD457 pKa = 3.21LGTTKK462 pKa = 10.93DD463 pKa = 3.25HH464 pKa = 5.83TTADD468 pKa = 3.65LRR470 pKa = 11.84TTVYY474 pKa = 10.85SCTADD479 pKa = 3.24LGTTKK484 pKa = 10.55DD485 pKa = 3.65YY486 pKa = 7.7TTADD490 pKa = 3.49PGTTEE495 pKa = 5.71DD496 pKa = 4.26YY497 pKa = 7.59TTKK500 pKa = 10.55HH501 pKa = 6.26LNTHH505 pKa = 5.61SEE507 pKa = 4.26GTDD510 pKa = 2.84SVTAVKK516 pKa = 8.77PTGLLTPIGIILISLAATTAIVALFVGLGLIVVSIWPRR554 pKa = 11.84NVKK557 pKa = 10.17EE558 pKa = 3.79IMGMKK563 pKa = 10.17DD564 pKa = 3.12SEE566 pKa = 4.45EE567 pKa = 4.22EE568 pKa = 3.91EE569 pKa = 4.86GMGSS573 pKa = 3.24
Molecular weight: 61.87 kDa
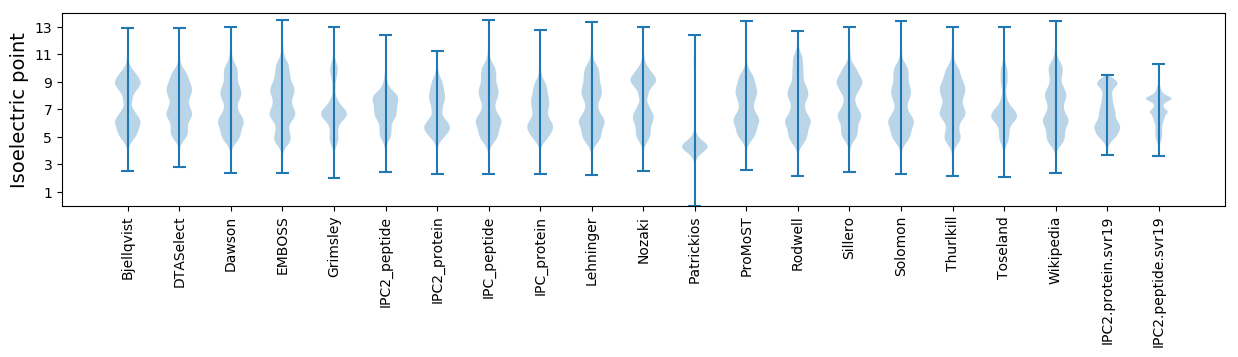
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5F5PPA3|A0A5F5PPA3_HORSE Immunity related GTPase Q OS=Equus caballus OX=9796 GN=IRGQ PE=3 SV=1
MM1 pKa = 7.1RR2 pKa = 11.84AKK4 pKa = 9.12WRR6 pKa = 11.84KK7 pKa = 9.1KK8 pKa = 9.32RR9 pKa = 11.84MRR11 pKa = 11.84RR12 pKa = 11.84LKK14 pKa = 10.08RR15 pKa = 11.84KK16 pKa = 8.21RR17 pKa = 11.84RR18 pKa = 11.84KK19 pKa = 8.46MRR21 pKa = 11.84QRR23 pKa = 11.84SKK25 pKa = 11.41
MM1 pKa = 7.1RR2 pKa = 11.84AKK4 pKa = 9.12WRR6 pKa = 11.84KK7 pKa = 9.1KK8 pKa = 9.32RR9 pKa = 11.84MRR11 pKa = 11.84RR12 pKa = 11.84LKK14 pKa = 10.08RR15 pKa = 11.84KK16 pKa = 8.21RR17 pKa = 11.84RR18 pKa = 11.84KK19 pKa = 8.46MRR21 pKa = 11.84QRR23 pKa = 11.84SKK25 pKa = 11.41
Molecular weight: 3.46 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
29288982 |
11 |
34311 |
658.3 |
73.32 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.05 ± 0.011 | 2.179 ± 0.01 |
4.804 ± 0.008 | 7.183 ± 0.017 |
3.565 ± 0.007 | 6.511 ± 0.014 |
2.542 ± 0.005 | 4.406 ± 0.011 |
5.834 ± 0.016 | 9.93 ± 0.018 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.114 ± 0.005 | 3.62 ± 0.008 |
6.313 ± 0.017 | 4.761 ± 0.013 |
5.763 ± 0.009 | 8.403 ± 0.015 |
5.276 ± 0.01 | 5.96 ± 0.013 |
1.206 ± 0.004 | 2.576 ± 0.006 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
