
Hubei sobemo-like virus 16
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 5.99
Get precalculated fractions of proteins
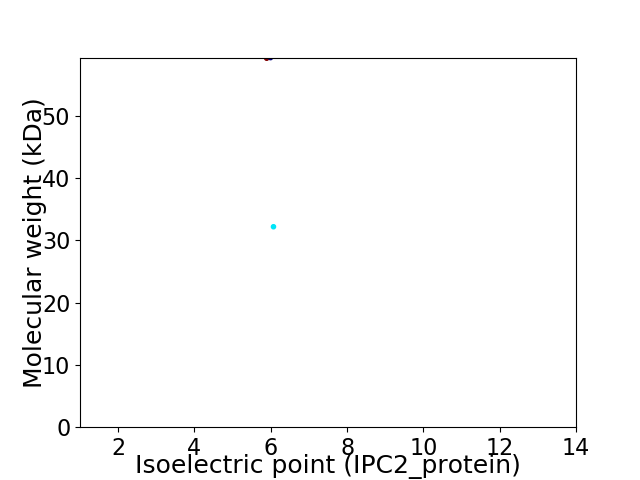
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KET4|A0A1L3KET4_9VIRU RdRP_1 domain-containing protein OS=Hubei sobemo-like virus 16 OX=1923201 PE=4 SV=1
MM1 pKa = 6.94PTVSIRR7 pKa = 11.84LPSLMDD13 pKa = 3.2VALLVVGTLYY23 pKa = 11.37LWMSWGVMTAPNAPAGVVGQNILTIVVFGFLYY55 pKa = 10.64LRR57 pKa = 11.84TGFASSGRR65 pKa = 11.84KK66 pKa = 8.74VVHH69 pKa = 5.6QVAKK73 pKa = 10.03SVSVLEE79 pKa = 4.06STIPGSHH86 pKa = 6.35IMEE89 pKa = 3.81VDD91 pKa = 4.55FKK93 pKa = 11.45NRR95 pKa = 11.84SLPKK99 pKa = 9.49CQTVVYY105 pKa = 10.47QEE107 pKa = 4.46TEE109 pKa = 3.76DD110 pKa = 4.41GEE112 pKa = 4.41YY113 pKa = 10.63HH114 pKa = 7.07LLGNVVRR121 pKa = 11.84IGDD124 pKa = 3.98FLVMPRR130 pKa = 11.84HH131 pKa = 5.84IADD134 pKa = 3.53VAKK137 pKa = 10.02EE138 pKa = 3.63LYY140 pKa = 9.56IAEE143 pKa = 4.37RR144 pKa = 11.84EE145 pKa = 4.28NFDD148 pKa = 4.04SVHH151 pKa = 4.65QVKK154 pKa = 10.26RR155 pKa = 11.84DD156 pKa = 3.27RR157 pKa = 11.84WEE159 pKa = 4.49DD160 pKa = 3.25LATDD164 pKa = 4.38LVYY167 pKa = 10.36MRR169 pKa = 11.84ASADD173 pKa = 3.77LLSTLQLSKK182 pKa = 11.36ASIGQLTTGIVASISTAAPSEE203 pKa = 4.17DD204 pKa = 3.4KK205 pKa = 10.98LLYY208 pKa = 9.49SCGVARR214 pKa = 11.84PSKK217 pKa = 10.65DD218 pKa = 2.61IFGLVEE224 pKa = 4.1YY225 pKa = 10.31KK226 pKa = 10.86GSTRR230 pKa = 11.84GGFSGAGLFVGTQLIGIHH248 pKa = 5.08YY249 pKa = 10.27QGTPSVNFSYY259 pKa = 10.29EE260 pKa = 3.56ASYY263 pKa = 10.45VKK265 pKa = 10.91AMLPRR270 pKa = 11.84EE271 pKa = 4.11EE272 pKa = 4.49ASDD275 pKa = 3.49WEE277 pKa = 4.36TAQWLEE283 pKa = 5.04DD284 pKa = 3.82IISKK288 pKa = 10.16KK289 pKa = 10.81GKK291 pKa = 9.35LKK293 pKa = 10.94AMRR296 pKa = 11.84DD297 pKa = 3.61PSDD300 pKa = 3.67PSDD303 pKa = 3.02WLVYY307 pKa = 10.76AGGKK311 pKa = 5.78YY312 pKa = 10.01HH313 pKa = 7.43RR314 pKa = 11.84IEE316 pKa = 5.52GDD318 pKa = 3.51VMSKK322 pKa = 10.95YY323 pKa = 10.53EE324 pKa = 4.19DD325 pKa = 3.87YY326 pKa = 11.77VDD328 pKa = 4.68FGYY331 pKa = 10.7DD332 pKa = 3.08EE333 pKa = 4.33PVARR337 pKa = 11.84RR338 pKa = 11.84EE339 pKa = 3.85MRR341 pKa = 11.84EE342 pKa = 3.38YY343 pKa = 10.98DD344 pKa = 3.33APHH347 pKa = 6.69YY348 pKa = 10.54SNQDD352 pKa = 2.97FRR354 pKa = 11.84VPPGLGNQEE363 pKa = 4.01ASHH366 pKa = 6.0AQPVCKK372 pKa = 10.22NLEE375 pKa = 3.81QLNILRR381 pKa = 11.84EE382 pKa = 4.16DD383 pKa = 3.71ADD385 pKa = 3.78NAKK388 pKa = 10.25QEE390 pKa = 4.08IDD392 pKa = 3.6DD393 pKa = 4.17LANVVDD399 pKa = 4.78EE400 pKa = 4.54VLHH403 pKa = 5.49AQKK406 pKa = 10.0ATVGSLEE413 pKa = 4.1EE414 pKa = 4.32LKK416 pKa = 10.76KK417 pKa = 10.35QQQEE421 pKa = 4.0LKK423 pKa = 10.65ALLRR427 pKa = 11.84KK428 pKa = 8.29LTQLPPTKK436 pKa = 10.05EE437 pKa = 3.92KK438 pKa = 10.57GVKK441 pKa = 8.29IHH443 pKa = 6.72SVEE446 pKa = 4.02EE447 pKa = 3.85QLNSNKK453 pKa = 10.36GEE455 pKa = 4.04IKK457 pKa = 10.53KK458 pKa = 10.0LANVPASSKK467 pKa = 10.68SPKK470 pKa = 8.28KK471 pKa = 8.76TKK473 pKa = 8.77TKK475 pKa = 9.58RR476 pKa = 11.84THH478 pKa = 6.22QDD480 pKa = 3.83LIQNLSPEE488 pKa = 4.38QIQAVCVINSNPEE501 pKa = 4.03LFRR504 pKa = 11.84VTQAALNNGEE514 pKa = 4.16VMQAILSRR522 pKa = 11.84LSQGRR527 pKa = 11.84NSISTSSSS535 pKa = 2.77
MM1 pKa = 6.94PTVSIRR7 pKa = 11.84LPSLMDD13 pKa = 3.2VALLVVGTLYY23 pKa = 11.37LWMSWGVMTAPNAPAGVVGQNILTIVVFGFLYY55 pKa = 10.64LRR57 pKa = 11.84TGFASSGRR65 pKa = 11.84KK66 pKa = 8.74VVHH69 pKa = 5.6QVAKK73 pKa = 10.03SVSVLEE79 pKa = 4.06STIPGSHH86 pKa = 6.35IMEE89 pKa = 3.81VDD91 pKa = 4.55FKK93 pKa = 11.45NRR95 pKa = 11.84SLPKK99 pKa = 9.49CQTVVYY105 pKa = 10.47QEE107 pKa = 4.46TEE109 pKa = 3.76DD110 pKa = 4.41GEE112 pKa = 4.41YY113 pKa = 10.63HH114 pKa = 7.07LLGNVVRR121 pKa = 11.84IGDD124 pKa = 3.98FLVMPRR130 pKa = 11.84HH131 pKa = 5.84IADD134 pKa = 3.53VAKK137 pKa = 10.02EE138 pKa = 3.63LYY140 pKa = 9.56IAEE143 pKa = 4.37RR144 pKa = 11.84EE145 pKa = 4.28NFDD148 pKa = 4.04SVHH151 pKa = 4.65QVKK154 pKa = 10.26RR155 pKa = 11.84DD156 pKa = 3.27RR157 pKa = 11.84WEE159 pKa = 4.49DD160 pKa = 3.25LATDD164 pKa = 4.38LVYY167 pKa = 10.36MRR169 pKa = 11.84ASADD173 pKa = 3.77LLSTLQLSKK182 pKa = 11.36ASIGQLTTGIVASISTAAPSEE203 pKa = 4.17DD204 pKa = 3.4KK205 pKa = 10.98LLYY208 pKa = 9.49SCGVARR214 pKa = 11.84PSKK217 pKa = 10.65DD218 pKa = 2.61IFGLVEE224 pKa = 4.1YY225 pKa = 10.31KK226 pKa = 10.86GSTRR230 pKa = 11.84GGFSGAGLFVGTQLIGIHH248 pKa = 5.08YY249 pKa = 10.27QGTPSVNFSYY259 pKa = 10.29EE260 pKa = 3.56ASYY263 pKa = 10.45VKK265 pKa = 10.91AMLPRR270 pKa = 11.84EE271 pKa = 4.11EE272 pKa = 4.49ASDD275 pKa = 3.49WEE277 pKa = 4.36TAQWLEE283 pKa = 5.04DD284 pKa = 3.82IISKK288 pKa = 10.16KK289 pKa = 10.81GKK291 pKa = 9.35LKK293 pKa = 10.94AMRR296 pKa = 11.84DD297 pKa = 3.61PSDD300 pKa = 3.67PSDD303 pKa = 3.02WLVYY307 pKa = 10.76AGGKK311 pKa = 5.78YY312 pKa = 10.01HH313 pKa = 7.43RR314 pKa = 11.84IEE316 pKa = 5.52GDD318 pKa = 3.51VMSKK322 pKa = 10.95YY323 pKa = 10.53EE324 pKa = 4.19DD325 pKa = 3.87YY326 pKa = 11.77VDD328 pKa = 4.68FGYY331 pKa = 10.7DD332 pKa = 3.08EE333 pKa = 4.33PVARR337 pKa = 11.84RR338 pKa = 11.84EE339 pKa = 3.85MRR341 pKa = 11.84EE342 pKa = 3.38YY343 pKa = 10.98DD344 pKa = 3.33APHH347 pKa = 6.69YY348 pKa = 10.54SNQDD352 pKa = 2.97FRR354 pKa = 11.84VPPGLGNQEE363 pKa = 4.01ASHH366 pKa = 6.0AQPVCKK372 pKa = 10.22NLEE375 pKa = 3.81QLNILRR381 pKa = 11.84EE382 pKa = 4.16DD383 pKa = 3.71ADD385 pKa = 3.78NAKK388 pKa = 10.25QEE390 pKa = 4.08IDD392 pKa = 3.6DD393 pKa = 4.17LANVVDD399 pKa = 4.78EE400 pKa = 4.54VLHH403 pKa = 5.49AQKK406 pKa = 10.0ATVGSLEE413 pKa = 4.1EE414 pKa = 4.32LKK416 pKa = 10.76KK417 pKa = 10.35QQQEE421 pKa = 4.0LKK423 pKa = 10.65ALLRR427 pKa = 11.84KK428 pKa = 8.29LTQLPPTKK436 pKa = 10.05EE437 pKa = 3.92KK438 pKa = 10.57GVKK441 pKa = 8.29IHH443 pKa = 6.72SVEE446 pKa = 4.02EE447 pKa = 3.85QLNSNKK453 pKa = 10.36GEE455 pKa = 4.04IKK457 pKa = 10.53KK458 pKa = 10.0LANVPASSKK467 pKa = 10.68SPKK470 pKa = 8.28KK471 pKa = 8.76TKK473 pKa = 8.77TKK475 pKa = 9.58RR476 pKa = 11.84THH478 pKa = 6.22QDD480 pKa = 3.83LIQNLSPEE488 pKa = 4.38QIQAVCVINSNPEE501 pKa = 4.03LFRR504 pKa = 11.84VTQAALNNGEE514 pKa = 4.16VMQAILSRR522 pKa = 11.84LSQGRR527 pKa = 11.84NSISTSSSS535 pKa = 2.77
Molecular weight: 59.26 kDa
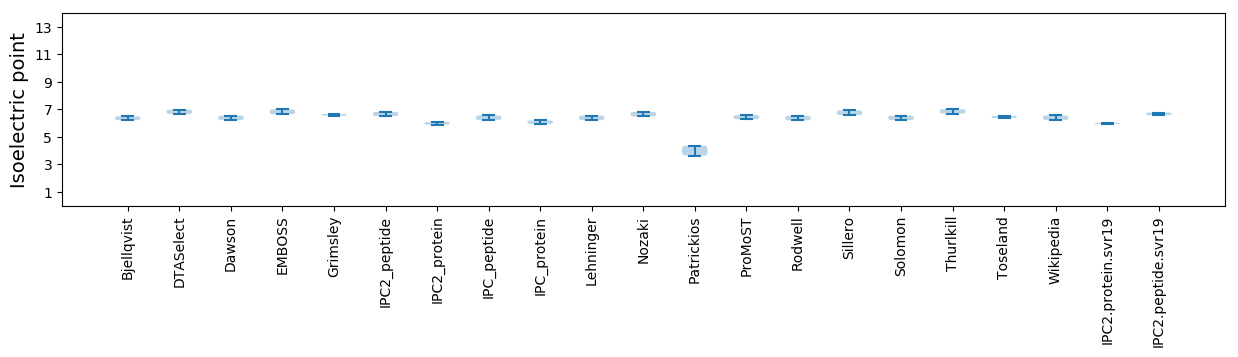
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KET4|A0A1L3KET4_9VIRU RdRP_1 domain-containing protein OS=Hubei sobemo-like virus 16 OX=1923201 PE=4 SV=1
MM1 pKa = 7.78LDD3 pKa = 2.85KK4 pKa = 10.53AYY6 pKa = 10.59RR7 pKa = 11.84IIHH10 pKa = 5.64GVSIIDD16 pKa = 3.58NLVARR21 pKa = 11.84LLFGNQFDD29 pKa = 4.21NLISNWSVLPSKK41 pKa = 10.92VGWTPGQGGYY51 pKa = 10.71KK52 pKa = 10.25LLAKK56 pKa = 9.97KK57 pKa = 10.45YY58 pKa = 8.65PYY60 pKa = 10.91SMSADD65 pKa = 3.07KK66 pKa = 11.1SAWDD70 pKa = 3.25WTMQPWCVDD79 pKa = 2.78LMIAMLRR86 pKa = 11.84NLGPQDD92 pKa = 3.12EE93 pKa = 4.79DD94 pKa = 4.02YY95 pKa = 11.31SRR97 pKa = 11.84RR98 pKa = 11.84VEE100 pKa = 3.92NHH102 pKa = 5.38LVALFGIGAKK112 pKa = 10.26FIAGNTVFTQRR123 pKa = 11.84VQGIMKK129 pKa = 9.96SGYY132 pKa = 10.33LGTIHH137 pKa = 7.03LNSILQTALHH147 pKa = 6.45ILAKK151 pKa = 10.01IRR153 pKa = 11.84LGQDD157 pKa = 2.94VEE159 pKa = 4.66PYY161 pKa = 9.04PDD163 pKa = 3.54SVGDD167 pKa = 3.77DD168 pKa = 3.77TIQRR172 pKa = 11.84PEE174 pKa = 4.56ADD176 pKa = 3.36PEE178 pKa = 4.73TYY180 pKa = 10.17FRR182 pKa = 11.84TLEE185 pKa = 4.3LGGCVIKK192 pKa = 10.64QYY194 pKa = 9.47EE195 pKa = 4.39TVKK198 pKa = 10.59GVDD201 pKa = 4.02FVGHH205 pKa = 6.97RR206 pKa = 11.84FNEE209 pKa = 4.42QQCLPNYY216 pKa = 9.71RR217 pKa = 11.84SKK219 pKa = 10.17HH220 pKa = 5.3CYY222 pKa = 9.75RR223 pKa = 11.84LFHH226 pKa = 7.86AEE228 pKa = 4.0DD229 pKa = 3.37QLVPEE234 pKa = 4.35TLEE237 pKa = 4.77GYY239 pKa = 10.09QYY241 pKa = 11.22LYY243 pKa = 10.52PHH245 pKa = 6.6DD246 pKa = 4.08TKK248 pKa = 10.79FLKK251 pKa = 10.5AIHH254 pKa = 6.57SMMVDD259 pKa = 3.66CDD261 pKa = 3.29AVDD264 pKa = 3.73RR265 pKa = 11.84MRR267 pKa = 11.84SPSYY271 pKa = 10.21LRR273 pKa = 11.84YY274 pKa = 8.69WYY276 pKa = 9.94DD277 pKa = 3.33YY278 pKa = 11.6AVV280 pKa = 3.16
MM1 pKa = 7.78LDD3 pKa = 2.85KK4 pKa = 10.53AYY6 pKa = 10.59RR7 pKa = 11.84IIHH10 pKa = 5.64GVSIIDD16 pKa = 3.58NLVARR21 pKa = 11.84LLFGNQFDD29 pKa = 4.21NLISNWSVLPSKK41 pKa = 10.92VGWTPGQGGYY51 pKa = 10.71KK52 pKa = 10.25LLAKK56 pKa = 9.97KK57 pKa = 10.45YY58 pKa = 8.65PYY60 pKa = 10.91SMSADD65 pKa = 3.07KK66 pKa = 11.1SAWDD70 pKa = 3.25WTMQPWCVDD79 pKa = 2.78LMIAMLRR86 pKa = 11.84NLGPQDD92 pKa = 3.12EE93 pKa = 4.79DD94 pKa = 4.02YY95 pKa = 11.31SRR97 pKa = 11.84RR98 pKa = 11.84VEE100 pKa = 3.92NHH102 pKa = 5.38LVALFGIGAKK112 pKa = 10.26FIAGNTVFTQRR123 pKa = 11.84VQGIMKK129 pKa = 9.96SGYY132 pKa = 10.33LGTIHH137 pKa = 7.03LNSILQTALHH147 pKa = 6.45ILAKK151 pKa = 10.01IRR153 pKa = 11.84LGQDD157 pKa = 2.94VEE159 pKa = 4.66PYY161 pKa = 9.04PDD163 pKa = 3.54SVGDD167 pKa = 3.77DD168 pKa = 3.77TIQRR172 pKa = 11.84PEE174 pKa = 4.56ADD176 pKa = 3.36PEE178 pKa = 4.73TYY180 pKa = 10.17FRR182 pKa = 11.84TLEE185 pKa = 4.3LGGCVIKK192 pKa = 10.64QYY194 pKa = 9.47EE195 pKa = 4.39TVKK198 pKa = 10.59GVDD201 pKa = 4.02FVGHH205 pKa = 6.97RR206 pKa = 11.84FNEE209 pKa = 4.42QQCLPNYY216 pKa = 9.71RR217 pKa = 11.84SKK219 pKa = 10.17HH220 pKa = 5.3CYY222 pKa = 9.75RR223 pKa = 11.84LFHH226 pKa = 7.86AEE228 pKa = 4.0DD229 pKa = 3.37QLVPEE234 pKa = 4.35TLEE237 pKa = 4.77GYY239 pKa = 10.09QYY241 pKa = 11.22LYY243 pKa = 10.52PHH245 pKa = 6.6DD246 pKa = 4.08TKK248 pKa = 10.79FLKK251 pKa = 10.5AIHH254 pKa = 6.57SMMVDD259 pKa = 3.66CDD261 pKa = 3.29AVDD264 pKa = 3.73RR265 pKa = 11.84MRR267 pKa = 11.84SPSYY271 pKa = 10.21LRR273 pKa = 11.84YY274 pKa = 8.69WYY276 pKa = 9.94DD277 pKa = 3.33YY278 pKa = 11.6AVV280 pKa = 3.16
Molecular weight: 32.19 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
815 |
280 |
535 |
407.5 |
45.72 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.871 ± 0.662 | 1.104 ± 0.39 |
6.012 ± 0.647 | 5.767 ± 1.052 |
2.822 ± 0.429 | 6.748 ± 0.226 |
2.577 ± 0.365 | 5.153 ± 0.321 |
6.012 ± 0.579 | 9.816 ± 0.105 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.577 ± 0.365 | 3.804 ± 0.133 |
4.54 ± 0.059 | 5.153 ± 0.088 |
4.908 ± 0.257 | 7.607 ± 1.287 |
4.54 ± 0.145 | 8.098 ± 0.751 |
1.472 ± 0.384 | 4.417 ± 1.151 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
