
Lake Sarah-associated circular virus-16
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 7.86
Get precalculated fractions of proteins
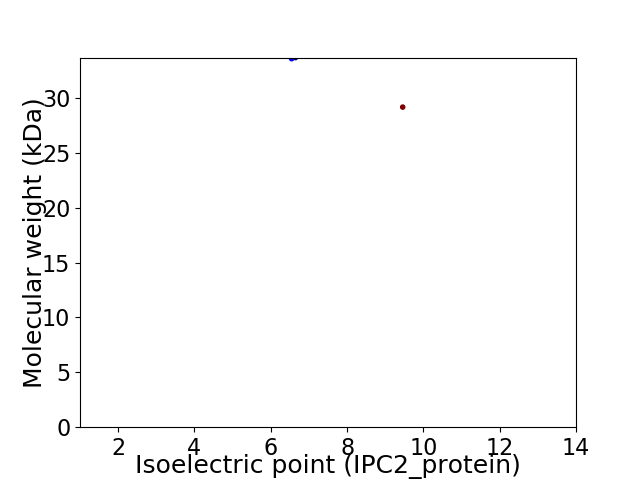
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A126GA49|A0A126GA49_9VIRU Coat protein OS=Lake Sarah-associated circular virus-16 OX=1685742 PE=4 SV=1
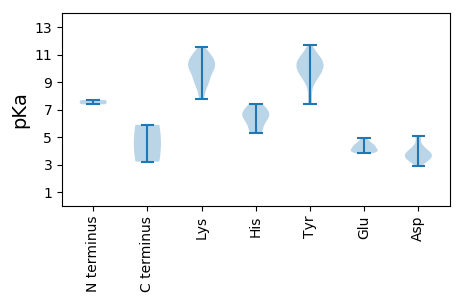
MM1 pKa = 7.7ANYY4 pKa = 9.44TDD6 pKa = 4.29ADD8 pKa = 3.57EE9 pKa = 4.83HH10 pKa = 6.69HH11 pKa = 6.89FLNTRR16 pKa = 11.84TKK18 pKa = 9.27AVHH21 pKa = 5.29FVSYY25 pKa = 7.65EE26 pKa = 3.98TSNEE30 pKa = 4.07TVEE33 pKa = 4.63RR34 pKa = 11.84LASLKK39 pKa = 9.25TDD41 pKa = 3.1YY42 pKa = 10.7MIIGDD47 pKa = 4.17EE48 pKa = 4.25HH49 pKa = 7.22CPTTGRR55 pKa = 11.84HH56 pKa = 5.74HH57 pKa = 6.47LQCYY61 pKa = 10.06LYY63 pKa = 8.99TKK65 pKa = 10.89NKK67 pKa = 7.78ITLSTIHH74 pKa = 6.78KK75 pKa = 10.34ACLKK79 pKa = 9.68KK80 pKa = 10.77CKK82 pKa = 10.29LFIAKK87 pKa = 8.02GTAEE91 pKa = 4.05QNQTYY96 pKa = 9.99CSKK99 pKa = 11.06EE100 pKa = 3.83KK101 pKa = 10.3ILYY104 pKa = 9.9EE105 pKa = 4.5SGTLPSGQGKK115 pKa = 7.84RR116 pKa = 11.84TDD118 pKa = 3.86LDD120 pKa = 3.49TTRR123 pKa = 11.84ANLLAGGTLRR133 pKa = 11.84DD134 pKa = 3.79VVLEE138 pKa = 3.96ATSYY142 pKa = 11.02QSVKK146 pKa = 9.9MAEE149 pKa = 4.24QILKK153 pKa = 8.87YY154 pKa = 10.59HH155 pKa = 6.22EE156 pKa = 4.68IKK158 pKa = 10.83RR159 pKa = 11.84NWKK162 pKa = 9.68PIVKK166 pKa = 8.73WFWGATGTGKK176 pKa = 10.58SLTAYY181 pKa = 10.54DD182 pKa = 4.92EE183 pKa = 4.3LDD185 pKa = 3.59KK186 pKa = 11.57DD187 pKa = 3.9DD188 pKa = 5.06TYY190 pKa = 11.73TAMSTGKK197 pKa = 8.6WWDD200 pKa = 3.61GYY202 pKa = 10.26DD203 pKa = 3.16AHH205 pKa = 7.43SCVVIDD211 pKa = 4.41DD212 pKa = 3.87MRR214 pKa = 11.84RR215 pKa = 11.84DD216 pKa = 3.4FCKK219 pKa = 9.89FHH221 pKa = 6.68EE222 pKa = 4.4LLRR225 pKa = 11.84YY226 pKa = 9.4LDD228 pKa = 4.34RR229 pKa = 11.84YY230 pKa = 7.4PTIVEE235 pKa = 4.4TKK237 pKa = 9.57GGTRR241 pKa = 11.84QFLAKK246 pKa = 10.19QIIITSCYY254 pKa = 9.96APEE257 pKa = 4.4DD258 pKa = 3.69MYY260 pKa = 10.55EE261 pKa = 4.0TRR263 pKa = 11.84EE264 pKa = 4.96DD265 pKa = 3.27IQQLLRR271 pKa = 11.84RR272 pKa = 11.84IDD274 pKa = 3.57EE275 pKa = 4.12VRR277 pKa = 11.84EE278 pKa = 4.04FTAEE282 pKa = 3.92SLGKK286 pKa = 10.19INSINN291 pKa = 3.21
MM1 pKa = 7.7ANYY4 pKa = 9.44TDD6 pKa = 4.29ADD8 pKa = 3.57EE9 pKa = 4.83HH10 pKa = 6.69HH11 pKa = 6.89FLNTRR16 pKa = 11.84TKK18 pKa = 9.27AVHH21 pKa = 5.29FVSYY25 pKa = 7.65EE26 pKa = 3.98TSNEE30 pKa = 4.07TVEE33 pKa = 4.63RR34 pKa = 11.84LASLKK39 pKa = 9.25TDD41 pKa = 3.1YY42 pKa = 10.7MIIGDD47 pKa = 4.17EE48 pKa = 4.25HH49 pKa = 7.22CPTTGRR55 pKa = 11.84HH56 pKa = 5.74HH57 pKa = 6.47LQCYY61 pKa = 10.06LYY63 pKa = 8.99TKK65 pKa = 10.89NKK67 pKa = 7.78ITLSTIHH74 pKa = 6.78KK75 pKa = 10.34ACLKK79 pKa = 9.68KK80 pKa = 10.77CKK82 pKa = 10.29LFIAKK87 pKa = 8.02GTAEE91 pKa = 4.05QNQTYY96 pKa = 9.99CSKK99 pKa = 11.06EE100 pKa = 3.83KK101 pKa = 10.3ILYY104 pKa = 9.9EE105 pKa = 4.5SGTLPSGQGKK115 pKa = 7.84RR116 pKa = 11.84TDD118 pKa = 3.86LDD120 pKa = 3.49TTRR123 pKa = 11.84ANLLAGGTLRR133 pKa = 11.84DD134 pKa = 3.79VVLEE138 pKa = 3.96ATSYY142 pKa = 11.02QSVKK146 pKa = 9.9MAEE149 pKa = 4.24QILKK153 pKa = 8.87YY154 pKa = 10.59HH155 pKa = 6.22EE156 pKa = 4.68IKK158 pKa = 10.83RR159 pKa = 11.84NWKK162 pKa = 9.68PIVKK166 pKa = 8.73WFWGATGTGKK176 pKa = 10.58SLTAYY181 pKa = 10.54DD182 pKa = 4.92EE183 pKa = 4.3LDD185 pKa = 3.59KK186 pKa = 11.57DD187 pKa = 3.9DD188 pKa = 5.06TYY190 pKa = 11.73TAMSTGKK197 pKa = 8.6WWDD200 pKa = 3.61GYY202 pKa = 10.26DD203 pKa = 3.16AHH205 pKa = 7.43SCVVIDD211 pKa = 4.41DD212 pKa = 3.87MRR214 pKa = 11.84RR215 pKa = 11.84DD216 pKa = 3.4FCKK219 pKa = 9.89FHH221 pKa = 6.68EE222 pKa = 4.4LLRR225 pKa = 11.84YY226 pKa = 9.4LDD228 pKa = 4.34RR229 pKa = 11.84YY230 pKa = 7.4PTIVEE235 pKa = 4.4TKK237 pKa = 9.57GGTRR241 pKa = 11.84QFLAKK246 pKa = 10.19QIIITSCYY254 pKa = 9.96APEE257 pKa = 4.4DD258 pKa = 3.69MYY260 pKa = 10.55EE261 pKa = 4.0TRR263 pKa = 11.84EE264 pKa = 4.96DD265 pKa = 3.27IQQLLRR271 pKa = 11.84RR272 pKa = 11.84IDD274 pKa = 3.57EE275 pKa = 4.12VRR277 pKa = 11.84EE278 pKa = 4.04FTAEE282 pKa = 3.92SLGKK286 pKa = 10.19INSINN291 pKa = 3.21
Molecular weight: 33.56 kDa
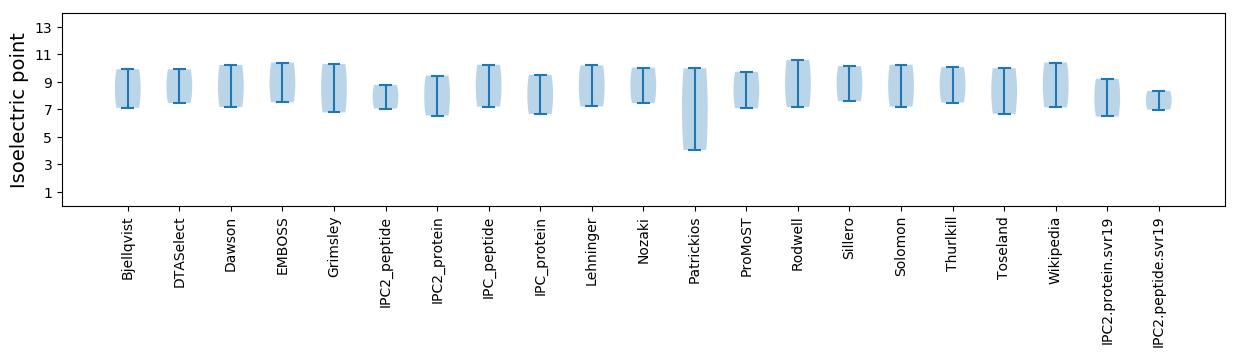
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A126GA49|A0A126GA49_9VIRU Coat protein OS=Lake Sarah-associated circular virus-16 OX=1685742 PE=4 SV=1
MM1 pKa = 7.38PRR3 pKa = 11.84STKK6 pKa = 10.12RR7 pKa = 11.84ATSRR11 pKa = 11.84RR12 pKa = 11.84PKK14 pKa = 10.02RR15 pKa = 11.84FARR18 pKa = 11.84RR19 pKa = 11.84KK20 pKa = 9.63AGRR23 pKa = 11.84KK24 pKa = 9.05SSGSQLVSKK33 pKa = 11.0AMLYY37 pKa = 10.32KK38 pKa = 10.73AIARR42 pKa = 11.84NLEE45 pKa = 3.87VKK47 pKa = 9.1MANIGFNYY55 pKa = 9.29TSQSSTLTSLATNFYY70 pKa = 10.86CPLPPVNLGIGSDD83 pKa = 3.49QRR85 pKa = 11.84IGDD88 pKa = 4.11KK89 pKa = 10.31IRR91 pKa = 11.84PLKK94 pKa = 10.56LVIRR98 pKa = 11.84GYY100 pKa = 10.28ISLVGDD106 pKa = 3.49VDD108 pKa = 3.96SSSRR112 pKa = 11.84ALAGRR117 pKa = 11.84LFCGLDD123 pKa = 3.19KK124 pKa = 11.21AILSKK129 pKa = 10.15TDD131 pKa = 3.82FSSLNINILDD141 pKa = 3.83NGGTSTTYY149 pKa = 10.69GGNLFGQCALKK160 pKa = 8.74NTQMYY165 pKa = 9.52RR166 pKa = 11.84WYY168 pKa = 10.5ADD170 pKa = 2.92KK171 pKa = 11.11KK172 pKa = 10.79FFLTKK177 pKa = 9.03PTGIYY182 pKa = 9.87QSAAIGGGVSSTPRR196 pKa = 11.84AQDD199 pKa = 3.3LFRR202 pKa = 11.84EE203 pKa = 4.42FTITLTKK210 pKa = 10.31KK211 pKa = 9.92HH212 pKa = 6.18LPAQFFYY219 pKa = 11.17DD220 pKa = 3.49STEE223 pKa = 3.89NATFPVNFCPFIALGYY239 pKa = 8.42CDD241 pKa = 4.96PMTSGIAGNGFMGMTYY257 pKa = 9.48TAVLHH262 pKa = 5.36YY263 pKa = 9.92TDD265 pKa = 3.93AA266 pKa = 5.89
MM1 pKa = 7.38PRR3 pKa = 11.84STKK6 pKa = 10.12RR7 pKa = 11.84ATSRR11 pKa = 11.84RR12 pKa = 11.84PKK14 pKa = 10.02RR15 pKa = 11.84FARR18 pKa = 11.84RR19 pKa = 11.84KK20 pKa = 9.63AGRR23 pKa = 11.84KK24 pKa = 9.05SSGSQLVSKK33 pKa = 11.0AMLYY37 pKa = 10.32KK38 pKa = 10.73AIARR42 pKa = 11.84NLEE45 pKa = 3.87VKK47 pKa = 9.1MANIGFNYY55 pKa = 9.29TSQSSTLTSLATNFYY70 pKa = 10.86CPLPPVNLGIGSDD83 pKa = 3.49QRR85 pKa = 11.84IGDD88 pKa = 4.11KK89 pKa = 10.31IRR91 pKa = 11.84PLKK94 pKa = 10.56LVIRR98 pKa = 11.84GYY100 pKa = 10.28ISLVGDD106 pKa = 3.49VDD108 pKa = 3.96SSSRR112 pKa = 11.84ALAGRR117 pKa = 11.84LFCGLDD123 pKa = 3.19KK124 pKa = 11.21AILSKK129 pKa = 10.15TDD131 pKa = 3.82FSSLNINILDD141 pKa = 3.83NGGTSTTYY149 pKa = 10.69GGNLFGQCALKK160 pKa = 8.74NTQMYY165 pKa = 9.52RR166 pKa = 11.84WYY168 pKa = 10.5ADD170 pKa = 2.92KK171 pKa = 11.11KK172 pKa = 10.79FFLTKK177 pKa = 9.03PTGIYY182 pKa = 9.87QSAAIGGGVSSTPRR196 pKa = 11.84AQDD199 pKa = 3.3LFRR202 pKa = 11.84EE203 pKa = 4.42FTITLTKK210 pKa = 10.31KK211 pKa = 9.92HH212 pKa = 6.18LPAQFFYY219 pKa = 11.17DD220 pKa = 3.49STEE223 pKa = 3.89NATFPVNFCPFIALGYY239 pKa = 8.42CDD241 pKa = 4.96PMTSGIAGNGFMGMTYY257 pKa = 9.48TAVLHH262 pKa = 5.36YY263 pKa = 9.92TDD265 pKa = 3.93AA266 pKa = 5.89
Molecular weight: 29.16 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
557 |
266 |
291 |
278.5 |
31.36 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.181 ± 0.738 | 2.334 ± 0.308 |
5.745 ± 0.835 | 4.129 ± 2.032 |
4.309 ± 1.155 | 7.181 ± 1.247 |
2.154 ± 0.95 | 5.925 ± 0.193 |
7.361 ± 0.657 | 8.797 ± 0.153 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.334 ± 0.202 | 3.95 ± 0.635 |
3.052 ± 0.988 | 3.232 ± 0.152 |
5.925 ± 0.316 | 7.002 ± 1.368 |
9.695 ± 0.71 | 3.591 ± 0.14 |
1.077 ± 0.475 | 5.027 ± 0.349 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
