
Epulopiscium sp. AS2M-Bin001
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Lachnospiraceae; Epulopiscium; unclassified Epulopiscium
Average proteome isoelectric point is 6.05
Get precalculated fractions of proteins
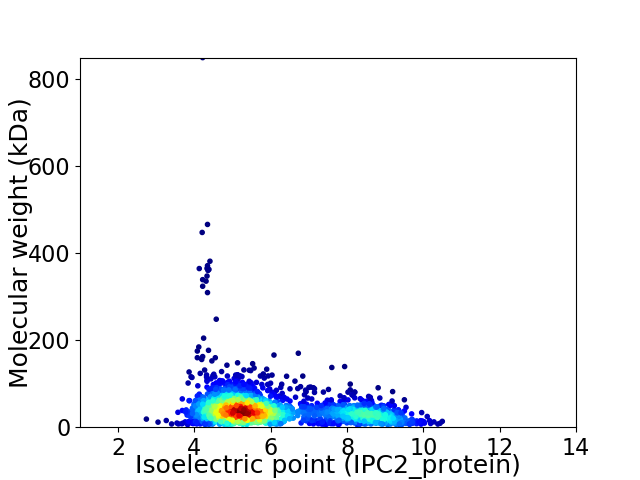
Virtual 2D-PAGE plot for 2165 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
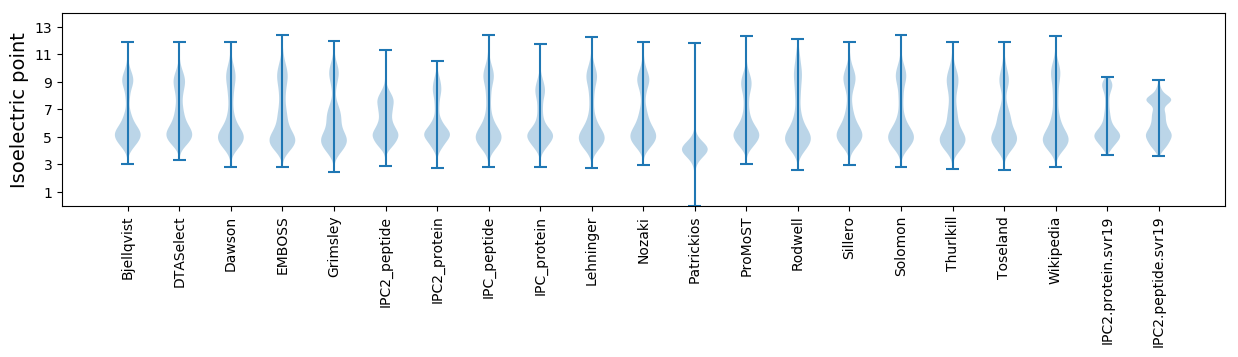
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1S9CFF4|A0A1S9CFF4_9FIRM Uncharacterized protein OS=Epulopiscium sp. AS2M-Bin001 OX=1764958 GN=ATN31_10760 PE=4 SV=1
MM1 pKa = 7.76KK2 pKa = 10.15KK3 pKa = 9.46ICLLASLFLTMSTNILADD21 pKa = 3.13SHH23 pKa = 7.22LFEE26 pKa = 4.64RR27 pKa = 11.84TATAIVADD35 pKa = 4.31YY36 pKa = 11.67VNVDD40 pKa = 3.88PKK42 pKa = 11.11NITISEE48 pKa = 4.19INSTDD53 pKa = 3.57DD54 pKa = 3.23QSTLHH59 pKa = 7.56IEE61 pKa = 4.41FFSPVIFSGFSFYY74 pKa = 11.09QLFKK78 pKa = 10.4CTLDD82 pKa = 3.5AQTGEE87 pKa = 4.29ILSSNLGANGYY98 pKa = 8.49ATEE101 pKa = 4.65LEE103 pKa = 4.54VPITPIQIEE112 pKa = 4.39TSNKK116 pKa = 8.37SHH118 pKa = 6.73SYY120 pKa = 9.24WEE122 pKa = 4.16SLVDD126 pKa = 4.35LNEE129 pKa = 4.03ALEE132 pKa = 4.37PCLEE136 pKa = 4.02GLEE139 pKa = 3.98WDD141 pKa = 4.34DD142 pKa = 5.94FINSTLADD150 pKa = 4.63FYY152 pKa = 11.36EE153 pKa = 4.39LEE155 pKa = 4.12IDD157 pKa = 4.86FKK159 pKa = 11.22QLVYY163 pKa = 10.92SHH165 pKa = 7.03GIEE168 pKa = 4.14DD169 pKa = 5.7LIYY172 pKa = 10.89HH173 pKa = 7.12LDD175 pKa = 3.34IKK177 pKa = 10.6DD178 pKa = 3.02WHH180 pKa = 6.25EE181 pKa = 4.62FISILEE187 pKa = 4.05DD188 pKa = 4.1EE189 pKa = 4.7IDD191 pKa = 3.43IVLDD195 pKa = 3.48NATSSLYY202 pKa = 10.51SDD204 pKa = 4.41YY205 pKa = 11.31NITQPEE211 pKa = 4.52LIEE214 pKa = 4.1TFEE217 pKa = 4.44FEE219 pKa = 3.98LTAITNEE226 pKa = 3.93WLDD229 pKa = 3.3NWKK232 pKa = 10.6SEE234 pKa = 3.91WGPKK238 pKa = 10.08LGVEE242 pKa = 5.22FKK244 pKa = 10.4MSYY247 pKa = 11.39DD248 pKa = 3.32NTDD251 pKa = 3.33INWYY255 pKa = 9.87NISDD259 pKa = 3.96NLDD262 pKa = 3.1QGLEE266 pKa = 4.07EE267 pKa = 6.03FSEE270 pKa = 4.5EE271 pKa = 3.97WQAGWEE277 pKa = 4.07NTLDD281 pKa = 3.02IWQDD285 pKa = 2.76ISDD288 pKa = 3.58NWKK291 pKa = 10.66SFWDD295 pKa = 3.62SLDD298 pKa = 3.4FF299 pKa = 5.13
MM1 pKa = 7.76KK2 pKa = 10.15KK3 pKa = 9.46ICLLASLFLTMSTNILADD21 pKa = 3.13SHH23 pKa = 7.22LFEE26 pKa = 4.64RR27 pKa = 11.84TATAIVADD35 pKa = 4.31YY36 pKa = 11.67VNVDD40 pKa = 3.88PKK42 pKa = 11.11NITISEE48 pKa = 4.19INSTDD53 pKa = 3.57DD54 pKa = 3.23QSTLHH59 pKa = 7.56IEE61 pKa = 4.41FFSPVIFSGFSFYY74 pKa = 11.09QLFKK78 pKa = 10.4CTLDD82 pKa = 3.5AQTGEE87 pKa = 4.29ILSSNLGANGYY98 pKa = 8.49ATEE101 pKa = 4.65LEE103 pKa = 4.54VPITPIQIEE112 pKa = 4.39TSNKK116 pKa = 8.37SHH118 pKa = 6.73SYY120 pKa = 9.24WEE122 pKa = 4.16SLVDD126 pKa = 4.35LNEE129 pKa = 4.03ALEE132 pKa = 4.37PCLEE136 pKa = 4.02GLEE139 pKa = 3.98WDD141 pKa = 4.34DD142 pKa = 5.94FINSTLADD150 pKa = 4.63FYY152 pKa = 11.36EE153 pKa = 4.39LEE155 pKa = 4.12IDD157 pKa = 4.86FKK159 pKa = 11.22QLVYY163 pKa = 10.92SHH165 pKa = 7.03GIEE168 pKa = 4.14DD169 pKa = 5.7LIYY172 pKa = 10.89HH173 pKa = 7.12LDD175 pKa = 3.34IKK177 pKa = 10.6DD178 pKa = 3.02WHH180 pKa = 6.25EE181 pKa = 4.62FISILEE187 pKa = 4.05DD188 pKa = 4.1EE189 pKa = 4.7IDD191 pKa = 3.43IVLDD195 pKa = 3.48NATSSLYY202 pKa = 10.51SDD204 pKa = 4.41YY205 pKa = 11.31NITQPEE211 pKa = 4.52LIEE214 pKa = 4.1TFEE217 pKa = 4.44FEE219 pKa = 3.98LTAITNEE226 pKa = 3.93WLDD229 pKa = 3.3NWKK232 pKa = 10.6SEE234 pKa = 3.91WGPKK238 pKa = 10.08LGVEE242 pKa = 5.22FKK244 pKa = 10.4MSYY247 pKa = 11.39DD248 pKa = 3.32NTDD251 pKa = 3.33INWYY255 pKa = 9.87NISDD259 pKa = 3.96NLDD262 pKa = 3.1QGLEE266 pKa = 4.07EE267 pKa = 6.03FSEE270 pKa = 4.5EE271 pKa = 3.97WQAGWEE277 pKa = 4.07NTLDD281 pKa = 3.02IWQDD285 pKa = 2.76ISDD288 pKa = 3.58NWKK291 pKa = 10.66SFWDD295 pKa = 3.62SLDD298 pKa = 3.4FF299 pKa = 5.13
Molecular weight: 34.64 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1S9CD82|A0A1S9CD82_9FIRM V-type ATP synthase beta chain OS=Epulopiscium sp. AS2M-Bin001 OX=1764958 GN=atpB PE=3 SV=1
MM1 pKa = 7.99AKK3 pKa = 10.23RR4 pKa = 11.84SMKK7 pKa = 10.21VKK9 pKa = 9.1QQRR12 pKa = 11.84PAKK15 pKa = 9.97FSTQAYY21 pKa = 8.53SRR23 pKa = 11.84CKK25 pKa = 9.47ICGRR29 pKa = 11.84PHH31 pKa = 7.33AYY33 pKa = 8.2IRR35 pKa = 11.84KK36 pKa = 8.92YY37 pKa = 10.18GICRR41 pKa = 11.84ICFRR45 pKa = 11.84EE46 pKa = 3.97LAYY49 pKa = 9.84KK50 pKa = 10.19GQIPGVKK57 pKa = 9.25KK58 pKa = 10.9ASWW61 pKa = 3.03
MM1 pKa = 7.99AKK3 pKa = 10.23RR4 pKa = 11.84SMKK7 pKa = 10.21VKK9 pKa = 9.1QQRR12 pKa = 11.84PAKK15 pKa = 9.97FSTQAYY21 pKa = 8.53SRR23 pKa = 11.84CKK25 pKa = 9.47ICGRR29 pKa = 11.84PHH31 pKa = 7.33AYY33 pKa = 8.2IRR35 pKa = 11.84KK36 pKa = 8.92YY37 pKa = 10.18GICRR41 pKa = 11.84ICFRR45 pKa = 11.84EE46 pKa = 3.97LAYY49 pKa = 9.84KK50 pKa = 10.19GQIPGVKK57 pKa = 9.25KK58 pKa = 10.9ASWW61 pKa = 3.03
Molecular weight: 7.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
826884 |
37 |
7665 |
381.9 |
42.93 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.365 ± 0.057 | 1.025 ± 0.022 |
6.036 ± 0.052 | 6.962 ± 0.058 |
4.4 ± 0.039 | 6.101 ± 0.059 |
1.646 ± 0.021 | 9.448 ± 0.067 |
7.286 ± 0.054 | 9.162 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.532 ± 0.03 | 6.222 ± 0.051 |
3.392 ± 0.037 | 3.162 ± 0.03 |
3.023 ± 0.035 | 6.447 ± 0.054 |
5.817 ± 0.066 | 6.032 ± 0.042 |
0.819 ± 0.017 | 4.122 ± 0.042 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
