
Gokushovirinae GAIR4
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; Gokushovirinae; unclassified Gokushovirinae
Average proteome isoelectric point is 8.72
Get precalculated fractions of proteins
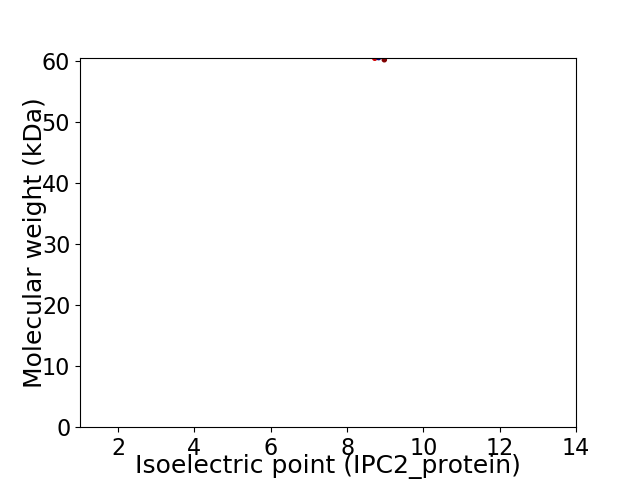
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0M5KZP2|A0A0M5KZP2_9VIRU Putative major capsid protein OS=Gokushovirinae GAIR4 OX=1701404 PE=3 SV=1
MM1 pKa = 7.49TMMHH5 pKa = 7.2KK6 pKa = 10.27NRR8 pKa = 11.84SVDD11 pKa = 3.13PHH13 pKa = 6.72RR14 pKa = 11.84FAMIPHH20 pKa = 6.93ADD22 pKa = 3.12IPRR25 pKa = 11.84AKK27 pKa = 10.09FDD29 pKa = 3.38RR30 pKa = 11.84QFTHH34 pKa = 6.1KK35 pKa = 7.96TTFDD39 pKa = 2.97AGYY42 pKa = 9.58LVPVYY47 pKa = 10.54VDD49 pKa = 3.45EE50 pKa = 4.56VLPGDD55 pKa = 3.93TFNLNMTAFARR66 pKa = 11.84LATPFFLLWIIWFWIVFFFFVPNRR90 pKa = 11.84LIWDD94 pKa = 3.67NWQKK98 pKa = 11.19FMVSSVTLVIRR109 pKa = 11.84LLCCSSTSFSRR120 pKa = 11.84RR121 pKa = 11.84WLCCWFFARR130 pKa = 11.84LYY132 pKa = 11.03GFTNCWSGWCWQYY145 pKa = 11.31YY146 pKa = 9.2FPLRR150 pKa = 11.84FFTRR154 pKa = 11.84AYY156 pKa = 10.45NLIWNEE162 pKa = 3.57WFRR165 pKa = 11.84DD166 pKa = 3.63QNLQNSVTVDD176 pKa = 3.12TGDD179 pKa = 4.08GPDD182 pKa = 3.83ASASTNYY189 pKa = 9.56TLLRR193 pKa = 11.84RR194 pKa = 11.84GKK196 pKa = 8.75RR197 pKa = 11.84HH198 pKa = 7.34DD199 pKa = 4.13YY200 pKa = 8.15FTSALPWPQKK210 pKa = 10.37GSSVTLPLGTSAPIKK225 pKa = 10.64SDD227 pKa = 3.11TTQVQFKK234 pKa = 10.79YY235 pKa = 10.74GATTFNSVSRR245 pKa = 11.84TDD247 pKa = 3.57VNVLGFNGAAVGAAGQAISFGANTGLYY274 pKa = 10.29ADD276 pKa = 5.23LSQATAATINQLRR289 pKa = 11.84QSFQIQKK296 pKa = 10.32LLEE299 pKa = 4.31RR300 pKa = 11.84DD301 pKa = 3.28ARR303 pKa = 11.84GGTRR307 pKa = 11.84YY308 pKa = 8.64TEE310 pKa = 4.53IIRR313 pKa = 11.84AHH315 pKa = 6.54FGVISPDD322 pKa = 3.07ARR324 pKa = 11.84LQRR327 pKa = 11.84PEE329 pKa = 3.87YY330 pKa = 10.14LGGGSTPVIINPIAQTSGTGVTGGSTPLANLAGVGTVLASGHH372 pKa = 6.33GFTQSFTEE380 pKa = 3.85HH381 pKa = 5.82GVIIGLVSIRR391 pKa = 11.84ADD393 pKa = 3.22LTYY396 pKa = 10.62QQGLHH401 pKa = 6.09RR402 pKa = 11.84MWSRR406 pKa = 11.84STRR409 pKa = 11.84YY410 pKa = 10.23DD411 pKa = 3.49FYY413 pKa = 11.54FPAFAMLGEE422 pKa = 4.0QPVYY426 pKa = 10.82NRR428 pKa = 11.84EE429 pKa = 3.98LYY431 pKa = 10.09IDD433 pKa = 4.42GSANDD438 pKa = 4.13GNVFGYY444 pKa = 7.33QEE446 pKa = 3.36RR447 pKa = 11.84WAEE450 pKa = 3.88YY451 pKa = 9.71RR452 pKa = 11.84YY453 pKa = 10.32KK454 pKa = 10.54PSQISSLFKK463 pKa = 9.81STSAGTIDD471 pKa = 3.86AWHH474 pKa = 7.28LAQKK478 pKa = 8.14FTSLPTLNSTFIQEE492 pKa = 4.33NPPVSRR498 pKa = 11.84VVAVGAAANGQQFIFDD514 pKa = 4.05SFFHH518 pKa = 6.06NVVARR523 pKa = 11.84PMPLYY528 pKa = 10.18SVPGMIDD535 pKa = 3.02HH536 pKa = 7.39FF537 pKa = 4.83
MM1 pKa = 7.49TMMHH5 pKa = 7.2KK6 pKa = 10.27NRR8 pKa = 11.84SVDD11 pKa = 3.13PHH13 pKa = 6.72RR14 pKa = 11.84FAMIPHH20 pKa = 6.93ADD22 pKa = 3.12IPRR25 pKa = 11.84AKK27 pKa = 10.09FDD29 pKa = 3.38RR30 pKa = 11.84QFTHH34 pKa = 6.1KK35 pKa = 7.96TTFDD39 pKa = 2.97AGYY42 pKa = 9.58LVPVYY47 pKa = 10.54VDD49 pKa = 3.45EE50 pKa = 4.56VLPGDD55 pKa = 3.93TFNLNMTAFARR66 pKa = 11.84LATPFFLLWIIWFWIVFFFFVPNRR90 pKa = 11.84LIWDD94 pKa = 3.67NWQKK98 pKa = 11.19FMVSSVTLVIRR109 pKa = 11.84LLCCSSTSFSRR120 pKa = 11.84RR121 pKa = 11.84WLCCWFFARR130 pKa = 11.84LYY132 pKa = 11.03GFTNCWSGWCWQYY145 pKa = 11.31YY146 pKa = 9.2FPLRR150 pKa = 11.84FFTRR154 pKa = 11.84AYY156 pKa = 10.45NLIWNEE162 pKa = 3.57WFRR165 pKa = 11.84DD166 pKa = 3.63QNLQNSVTVDD176 pKa = 3.12TGDD179 pKa = 4.08GPDD182 pKa = 3.83ASASTNYY189 pKa = 9.56TLLRR193 pKa = 11.84RR194 pKa = 11.84GKK196 pKa = 8.75RR197 pKa = 11.84HH198 pKa = 7.34DD199 pKa = 4.13YY200 pKa = 8.15FTSALPWPQKK210 pKa = 10.37GSSVTLPLGTSAPIKK225 pKa = 10.64SDD227 pKa = 3.11TTQVQFKK234 pKa = 10.79YY235 pKa = 10.74GATTFNSVSRR245 pKa = 11.84TDD247 pKa = 3.57VNVLGFNGAAVGAAGQAISFGANTGLYY274 pKa = 10.29ADD276 pKa = 5.23LSQATAATINQLRR289 pKa = 11.84QSFQIQKK296 pKa = 10.32LLEE299 pKa = 4.31RR300 pKa = 11.84DD301 pKa = 3.28ARR303 pKa = 11.84GGTRR307 pKa = 11.84YY308 pKa = 8.64TEE310 pKa = 4.53IIRR313 pKa = 11.84AHH315 pKa = 6.54FGVISPDD322 pKa = 3.07ARR324 pKa = 11.84LQRR327 pKa = 11.84PEE329 pKa = 3.87YY330 pKa = 10.14LGGGSTPVIINPIAQTSGTGVTGGSTPLANLAGVGTVLASGHH372 pKa = 6.33GFTQSFTEE380 pKa = 3.85HH381 pKa = 5.82GVIIGLVSIRR391 pKa = 11.84ADD393 pKa = 3.22LTYY396 pKa = 10.62QQGLHH401 pKa = 6.09RR402 pKa = 11.84MWSRR406 pKa = 11.84STRR409 pKa = 11.84YY410 pKa = 10.23DD411 pKa = 3.49FYY413 pKa = 11.54FPAFAMLGEE422 pKa = 4.0QPVYY426 pKa = 10.82NRR428 pKa = 11.84EE429 pKa = 3.98LYY431 pKa = 10.09IDD433 pKa = 4.42GSANDD438 pKa = 4.13GNVFGYY444 pKa = 7.33QEE446 pKa = 3.36RR447 pKa = 11.84WAEE450 pKa = 3.88YY451 pKa = 9.71RR452 pKa = 11.84YY453 pKa = 10.32KK454 pKa = 10.54PSQISSLFKK463 pKa = 9.81STSAGTIDD471 pKa = 3.86AWHH474 pKa = 7.28LAQKK478 pKa = 8.14FTSLPTLNSTFIQEE492 pKa = 4.33NPPVSRR498 pKa = 11.84VVAVGAAANGQQFIFDD514 pKa = 4.05SFFHH518 pKa = 6.06NVVARR523 pKa = 11.84PMPLYY528 pKa = 10.18SVPGMIDD535 pKa = 3.02HH536 pKa = 7.39FF537 pKa = 4.83
Molecular weight: 60.46 kDa
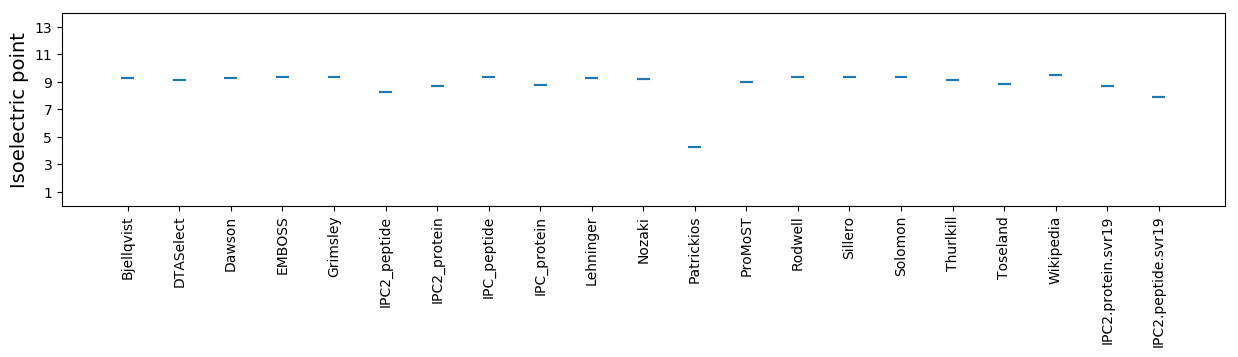
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0M5KZP2|A0A0M5KZP2_9VIRU Putative major capsid protein OS=Gokushovirinae GAIR4 OX=1701404 PE=3 SV=1
MM1 pKa = 7.49TMMHH5 pKa = 7.2KK6 pKa = 10.27NRR8 pKa = 11.84SVDD11 pKa = 3.13PHH13 pKa = 6.72RR14 pKa = 11.84FAMIPHH20 pKa = 6.93ADD22 pKa = 3.12IPRR25 pKa = 11.84AKK27 pKa = 10.09FDD29 pKa = 3.38RR30 pKa = 11.84QFTHH34 pKa = 6.1KK35 pKa = 7.96TTFDD39 pKa = 2.97AGYY42 pKa = 9.58LVPVYY47 pKa = 10.54VDD49 pKa = 3.45EE50 pKa = 4.56VLPGDD55 pKa = 3.93TFNLNMTAFARR66 pKa = 11.84LATPFFLLWIIWFWIVFFFFVPNRR90 pKa = 11.84LIWDD94 pKa = 3.67NWQKK98 pKa = 11.19FMVSSVTLVIRR109 pKa = 11.84LLCCSSTSFSRR120 pKa = 11.84RR121 pKa = 11.84WLCCWFFARR130 pKa = 11.84LYY132 pKa = 11.03GFTNCWSGWCWQYY145 pKa = 11.31YY146 pKa = 9.2FPLRR150 pKa = 11.84FFTRR154 pKa = 11.84AYY156 pKa = 10.45NLIWNEE162 pKa = 3.57WFRR165 pKa = 11.84DD166 pKa = 3.63QNLQNSVTVDD176 pKa = 3.12TGDD179 pKa = 4.08GPDD182 pKa = 3.83ASASTNYY189 pKa = 9.56TLLRR193 pKa = 11.84RR194 pKa = 11.84GKK196 pKa = 8.75RR197 pKa = 11.84HH198 pKa = 7.34DD199 pKa = 4.13YY200 pKa = 8.15FTSALPWPQKK210 pKa = 10.37GSSVTLPLGTSAPIKK225 pKa = 10.64SDD227 pKa = 3.11TTQVQFKK234 pKa = 10.79YY235 pKa = 10.74GATTFNSVSRR245 pKa = 11.84TDD247 pKa = 3.57VNVLGFNGAAVGAAGQAISFGANTGLYY274 pKa = 10.29ADD276 pKa = 5.23LSQATAATINQLRR289 pKa = 11.84QSFQIQKK296 pKa = 10.32LLEE299 pKa = 4.31RR300 pKa = 11.84DD301 pKa = 3.28ARR303 pKa = 11.84GGTRR307 pKa = 11.84YY308 pKa = 8.64TEE310 pKa = 4.53IIRR313 pKa = 11.84AHH315 pKa = 6.54FGVISPDD322 pKa = 3.07ARR324 pKa = 11.84LQRR327 pKa = 11.84PEE329 pKa = 3.87YY330 pKa = 10.14LGGGSTPVIINPIAQTSGTGVTGGSTPLANLAGVGTVLASGHH372 pKa = 6.33GFTQSFTEE380 pKa = 3.85HH381 pKa = 5.82GVIIGLVSIRR391 pKa = 11.84ADD393 pKa = 3.22LTYY396 pKa = 10.62QQGLHH401 pKa = 6.09RR402 pKa = 11.84MWSRR406 pKa = 11.84STRR409 pKa = 11.84YY410 pKa = 10.23DD411 pKa = 3.49FYY413 pKa = 11.54FPAFAMLGEE422 pKa = 4.0QPVYY426 pKa = 10.82NRR428 pKa = 11.84EE429 pKa = 3.98LYY431 pKa = 10.09IDD433 pKa = 4.42GSANDD438 pKa = 4.13GNVFGYY444 pKa = 7.33QEE446 pKa = 3.36RR447 pKa = 11.84WAEE450 pKa = 3.88YY451 pKa = 9.71RR452 pKa = 11.84YY453 pKa = 10.32KK454 pKa = 10.54PSQISSLFKK463 pKa = 9.81STSAGTIDD471 pKa = 3.86AWHH474 pKa = 7.28LAQKK478 pKa = 8.14FTSLPTLNSTFIQEE492 pKa = 4.33NPPVSRR498 pKa = 11.84VVAVGAAANGQQFIFDD514 pKa = 4.05SFFHH518 pKa = 6.06NVVARR523 pKa = 11.84PMPLYY528 pKa = 10.18SVPGMIDD535 pKa = 3.02HH536 pKa = 7.39FF537 pKa = 4.83
MM1 pKa = 7.49TMMHH5 pKa = 7.2KK6 pKa = 10.27NRR8 pKa = 11.84SVDD11 pKa = 3.13PHH13 pKa = 6.72RR14 pKa = 11.84FAMIPHH20 pKa = 6.93ADD22 pKa = 3.12IPRR25 pKa = 11.84AKK27 pKa = 10.09FDD29 pKa = 3.38RR30 pKa = 11.84QFTHH34 pKa = 6.1KK35 pKa = 7.96TTFDD39 pKa = 2.97AGYY42 pKa = 9.58LVPVYY47 pKa = 10.54VDD49 pKa = 3.45EE50 pKa = 4.56VLPGDD55 pKa = 3.93TFNLNMTAFARR66 pKa = 11.84LATPFFLLWIIWFWIVFFFFVPNRR90 pKa = 11.84LIWDD94 pKa = 3.67NWQKK98 pKa = 11.19FMVSSVTLVIRR109 pKa = 11.84LLCCSSTSFSRR120 pKa = 11.84RR121 pKa = 11.84WLCCWFFARR130 pKa = 11.84LYY132 pKa = 11.03GFTNCWSGWCWQYY145 pKa = 11.31YY146 pKa = 9.2FPLRR150 pKa = 11.84FFTRR154 pKa = 11.84AYY156 pKa = 10.45NLIWNEE162 pKa = 3.57WFRR165 pKa = 11.84DD166 pKa = 3.63QNLQNSVTVDD176 pKa = 3.12TGDD179 pKa = 4.08GPDD182 pKa = 3.83ASASTNYY189 pKa = 9.56TLLRR193 pKa = 11.84RR194 pKa = 11.84GKK196 pKa = 8.75RR197 pKa = 11.84HH198 pKa = 7.34DD199 pKa = 4.13YY200 pKa = 8.15FTSALPWPQKK210 pKa = 10.37GSSVTLPLGTSAPIKK225 pKa = 10.64SDD227 pKa = 3.11TTQVQFKK234 pKa = 10.79YY235 pKa = 10.74GATTFNSVSRR245 pKa = 11.84TDD247 pKa = 3.57VNVLGFNGAAVGAAGQAISFGANTGLYY274 pKa = 10.29ADD276 pKa = 5.23LSQATAATINQLRR289 pKa = 11.84QSFQIQKK296 pKa = 10.32LLEE299 pKa = 4.31RR300 pKa = 11.84DD301 pKa = 3.28ARR303 pKa = 11.84GGTRR307 pKa = 11.84YY308 pKa = 8.64TEE310 pKa = 4.53IIRR313 pKa = 11.84AHH315 pKa = 6.54FGVISPDD322 pKa = 3.07ARR324 pKa = 11.84LQRR327 pKa = 11.84PEE329 pKa = 3.87YY330 pKa = 10.14LGGGSTPVIINPIAQTSGTGVTGGSTPLANLAGVGTVLASGHH372 pKa = 6.33GFTQSFTEE380 pKa = 3.85HH381 pKa = 5.82GVIIGLVSIRR391 pKa = 11.84ADD393 pKa = 3.22LTYY396 pKa = 10.62QQGLHH401 pKa = 6.09RR402 pKa = 11.84MWSRR406 pKa = 11.84STRR409 pKa = 11.84YY410 pKa = 10.23DD411 pKa = 3.49FYY413 pKa = 11.54FPAFAMLGEE422 pKa = 4.0QPVYY426 pKa = 10.82NRR428 pKa = 11.84EE429 pKa = 3.98LYY431 pKa = 10.09IDD433 pKa = 4.42GSANDD438 pKa = 4.13GNVFGYY444 pKa = 7.33QEE446 pKa = 3.36RR447 pKa = 11.84WAEE450 pKa = 3.88YY451 pKa = 9.71RR452 pKa = 11.84YY453 pKa = 10.32KK454 pKa = 10.54PSQISSLFKK463 pKa = 9.81STSAGTIDD471 pKa = 3.86AWHH474 pKa = 7.28LAQKK478 pKa = 8.14FTSLPTLNSTFIQEE492 pKa = 4.33NPPVSRR498 pKa = 11.84VVAVGAAANGQQFIFDD514 pKa = 4.05SFFHH518 pKa = 6.06NVVARR523 pKa = 11.84PMPLYY528 pKa = 10.18SVPGMIDD535 pKa = 3.02HH536 pKa = 7.39FF537 pKa = 4.83
Molecular weight: 60.46 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
537 |
537 |
537 |
537.0 |
60.46 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.194 ± 0.0 | 1.117 ± 0.0 |
4.469 ± 0.0 | 2.048 ± 0.0 |
8.007 ± 0.0 | 7.635 ± 0.0 |
2.235 ± 0.0 | 5.028 ± 0.0 |
2.235 ± 0.0 | 7.635 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.862 ± 0.0 | 4.655 ± 0.0 |
5.028 ± 0.0 | 4.842 ± 0.0 |
6.145 ± 0.0 | 7.635 ± 0.0 |
8.007 ± 0.0 | 6.331 ± 0.0 |
2.98 ± 0.0 | 3.911 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
