
Penicillium steckii
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; Eurotiomycetes; Eurotiomycetidae; Eurotiales; Aspergillaceae; Penicillium
Average proteome isoelectric point is 6.26
Get precalculated fractions of proteins
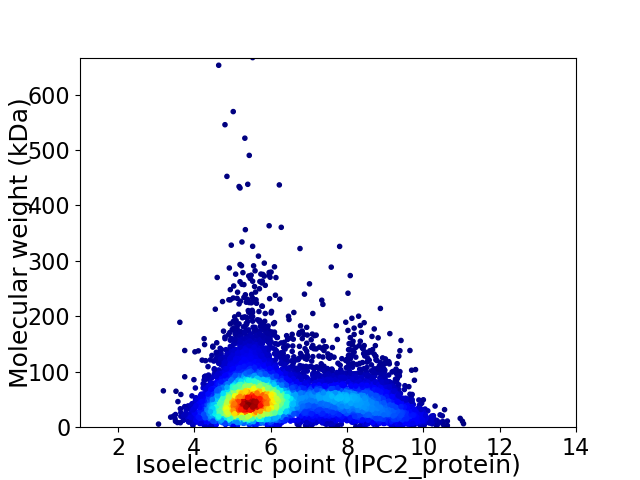
Virtual 2D-PAGE plot for 10355 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1V6TZC2|A0A1V6TZC2_9EURO Uncharacterized protein OS=Penicillium steckii OX=303698 GN=PENSTE_c001G06324 PE=3 SV=1
MM1 pKa = 7.11TLKK4 pKa = 10.77LQALLTAASALLINGVAIAQPTRR27 pKa = 11.84VIDD30 pKa = 3.45TDD32 pKa = 3.79FADD35 pKa = 3.84PCVIQTDD42 pKa = 3.45DD43 pKa = 3.81GYY45 pKa = 11.77YY46 pKa = 11.06AFATSGNGVNVQIASSSDD64 pKa = 3.32FASWEE69 pKa = 4.05LLSGNDD75 pKa = 3.6AMPGPFPSWVADD87 pKa = 4.29TPAIWAPDD95 pKa = 3.62VIQRR99 pKa = 11.84DD100 pKa = 3.64DD101 pKa = 3.78GTFVMYY107 pKa = 8.94FSAASADD114 pKa = 4.23DD115 pKa = 4.16DD116 pKa = 4.69SKK118 pKa = 11.39HH119 pKa = 6.26CVGAATSSSITGPYY133 pKa = 8.7TPEE136 pKa = 4.51DD137 pKa = 3.59NALACPLDD145 pKa = 3.6KK146 pKa = 11.22GGAIDD151 pKa = 5.27ADD153 pKa = 4.01GFKK156 pKa = 11.09DD157 pKa = 3.11GDD159 pKa = 3.8TYY161 pKa = 11.49YY162 pKa = 11.16VVYY165 pKa = 10.45KK166 pKa = 10.31IDD168 pKa = 3.86GNSIQSDD175 pKa = 4.11STPLMLQGLNSDD187 pKa = 3.39ATTANGDD194 pKa = 3.45PTQLLDD200 pKa = 5.26RR201 pKa = 11.84DD202 pKa = 4.25DD203 pKa = 3.89TDD205 pKa = 4.0GPLIEE210 pKa = 5.28APSLANVDD218 pKa = 3.23GTYY221 pKa = 10.95YY222 pKa = 11.08LSFSSNVYY230 pKa = 7.94STKK233 pKa = 10.75DD234 pKa = 3.31YY235 pKa = 11.21DD236 pKa = 3.75VSYY239 pKa = 9.31ATASSLTGPYY249 pKa = 9.17TKK251 pKa = 10.63ARR253 pKa = 11.84DD254 pKa = 3.5PDD256 pKa = 3.71APLLVSGDD264 pKa = 3.77PSNVGDD270 pKa = 4.0LGGPGGSDD278 pKa = 3.5FNADD282 pKa = 2.74GSKK285 pKa = 10.08IVFHH289 pKa = 6.81AFEE292 pKa = 5.02NGEE295 pKa = 4.05NMDD298 pKa = 3.9KK299 pKa = 10.9GRR301 pKa = 11.84AMWVADD307 pKa = 3.51ITCASGVITINN318 pKa = 3.16
MM1 pKa = 7.11TLKK4 pKa = 10.77LQALLTAASALLINGVAIAQPTRR27 pKa = 11.84VIDD30 pKa = 3.45TDD32 pKa = 3.79FADD35 pKa = 3.84PCVIQTDD42 pKa = 3.45DD43 pKa = 3.81GYY45 pKa = 11.77YY46 pKa = 11.06AFATSGNGVNVQIASSSDD64 pKa = 3.32FASWEE69 pKa = 4.05LLSGNDD75 pKa = 3.6AMPGPFPSWVADD87 pKa = 4.29TPAIWAPDD95 pKa = 3.62VIQRR99 pKa = 11.84DD100 pKa = 3.64DD101 pKa = 3.78GTFVMYY107 pKa = 8.94FSAASADD114 pKa = 4.23DD115 pKa = 4.16DD116 pKa = 4.69SKK118 pKa = 11.39HH119 pKa = 6.26CVGAATSSSITGPYY133 pKa = 8.7TPEE136 pKa = 4.51DD137 pKa = 3.59NALACPLDD145 pKa = 3.6KK146 pKa = 11.22GGAIDD151 pKa = 5.27ADD153 pKa = 4.01GFKK156 pKa = 11.09DD157 pKa = 3.11GDD159 pKa = 3.8TYY161 pKa = 11.49YY162 pKa = 11.16VVYY165 pKa = 10.45KK166 pKa = 10.31IDD168 pKa = 3.86GNSIQSDD175 pKa = 4.11STPLMLQGLNSDD187 pKa = 3.39ATTANGDD194 pKa = 3.45PTQLLDD200 pKa = 5.26RR201 pKa = 11.84DD202 pKa = 4.25DD203 pKa = 3.89TDD205 pKa = 4.0GPLIEE210 pKa = 5.28APSLANVDD218 pKa = 3.23GTYY221 pKa = 10.95YY222 pKa = 11.08LSFSSNVYY230 pKa = 7.94STKK233 pKa = 10.75DD234 pKa = 3.31YY235 pKa = 11.21DD236 pKa = 3.75VSYY239 pKa = 9.31ATASSLTGPYY249 pKa = 9.17TKK251 pKa = 10.63ARR253 pKa = 11.84DD254 pKa = 3.5PDD256 pKa = 3.71APLLVSGDD264 pKa = 3.77PSNVGDD270 pKa = 4.0LGGPGGSDD278 pKa = 3.5FNADD282 pKa = 2.74GSKK285 pKa = 10.08IVFHH289 pKa = 6.81AFEE292 pKa = 5.02NGEE295 pKa = 4.05NMDD298 pKa = 3.9KK299 pKa = 10.9GRR301 pKa = 11.84AMWVADD307 pKa = 3.51ITCASGVITINN318 pKa = 3.16
Molecular weight: 33.22 kDa
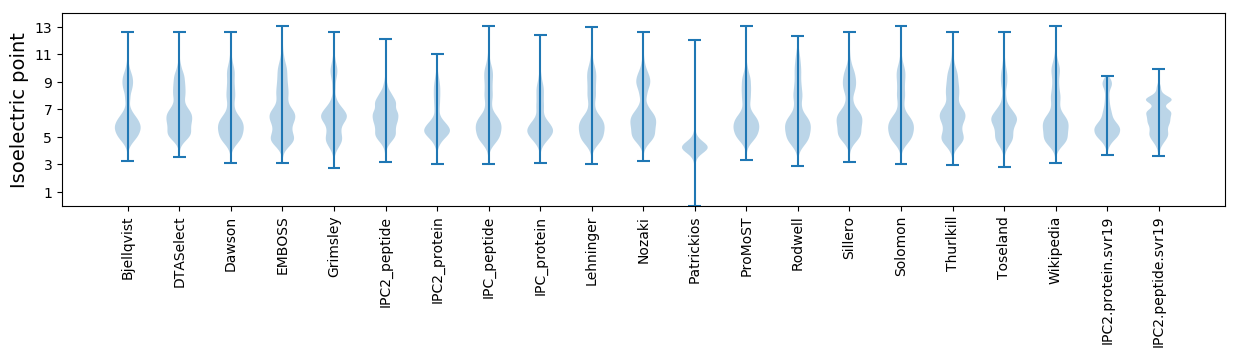
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1V6TP39|A0A1V6TP39_9EURO Copper homeostasis protein cutC homolog OS=Penicillium steckii OX=303698 GN=PENSTE_c004G02620 PE=3 SV=1
MM1 pKa = 7.6LCFRR5 pKa = 11.84CRR7 pKa = 11.84AVPSLRR13 pKa = 11.84ASVSTPRR20 pKa = 11.84ALMYY24 pKa = 10.24SASPFAVQSKK34 pKa = 10.38AITSAFPSARR44 pKa = 11.84PFSSTLLSTPVRR56 pKa = 11.84PQQTQMLSRR65 pKa = 11.84LPSTTLSATPSASSLLSQQSQQSRR89 pKa = 11.84AFSATAHH96 pKa = 6.38LGVKK100 pKa = 10.16RR101 pKa = 11.84NTFNPSRR108 pKa = 11.84RR109 pKa = 11.84VQKK112 pKa = 10.39RR113 pKa = 11.84RR114 pKa = 11.84SGFLARR120 pKa = 11.84NRR122 pKa = 11.84SQKK125 pKa = 10.12GRR127 pKa = 11.84LVLIRR132 pKa = 11.84RR133 pKa = 11.84RR134 pKa = 11.84LKK136 pKa = 10.43GRR138 pKa = 11.84KK139 pKa = 8.86AMSWW143 pKa = 3.05
MM1 pKa = 7.6LCFRR5 pKa = 11.84CRR7 pKa = 11.84AVPSLRR13 pKa = 11.84ASVSTPRR20 pKa = 11.84ALMYY24 pKa = 10.24SASPFAVQSKK34 pKa = 10.38AITSAFPSARR44 pKa = 11.84PFSSTLLSTPVRR56 pKa = 11.84PQQTQMLSRR65 pKa = 11.84LPSTTLSATPSASSLLSQQSQQSRR89 pKa = 11.84AFSATAHH96 pKa = 6.38LGVKK100 pKa = 10.16RR101 pKa = 11.84NTFNPSRR108 pKa = 11.84RR109 pKa = 11.84VQKK112 pKa = 10.39RR113 pKa = 11.84RR114 pKa = 11.84SGFLARR120 pKa = 11.84NRR122 pKa = 11.84SQKK125 pKa = 10.12GRR127 pKa = 11.84LVLIRR132 pKa = 11.84RR133 pKa = 11.84RR134 pKa = 11.84LKK136 pKa = 10.43GRR138 pKa = 11.84KK139 pKa = 8.86AMSWW143 pKa = 3.05
Molecular weight: 15.85 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5147105 |
8 |
6030 |
497.1 |
55.11 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.066 ± 0.019 | 1.241 ± 0.008 |
5.712 ± 0.017 | 6.165 ± 0.027 |
3.887 ± 0.015 | 6.786 ± 0.026 |
2.383 ± 0.01 | 5.261 ± 0.016 |
4.753 ± 0.022 | 8.9 ± 0.028 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.247 ± 0.008 | 3.946 ± 0.012 |
5.921 ± 0.027 | 4.084 ± 0.017 |
5.831 ± 0.02 | 8.666 ± 0.027 |
5.911 ± 0.021 | 5.927 ± 0.015 |
1.487 ± 0.009 | 2.825 ± 0.012 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
