
Alteromonas pelagimontana
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Alteromonadales; Alteromonadaceae; Alteromonas
Average proteome isoelectric point is 6.15
Get precalculated fractions of proteins
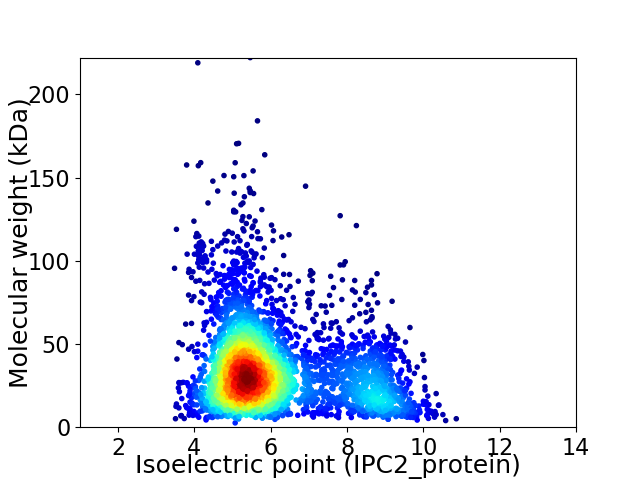
Virtual 2D-PAGE plot for 3617 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6M4MC23|A0A6M4MC23_9ALTE DUF1624 domain-containing protein OS=Alteromonas pelagimontana OX=1858656 GN=CA267_008035 PE=4 SV=1
MM1 pKa = 7.73RR2 pKa = 11.84KK3 pKa = 8.14ITLSSRR9 pKa = 11.84QLSKK13 pKa = 10.66IALASALAFCSSQSHH28 pKa = 5.84AAVGNLIWEE37 pKa = 4.7DD38 pKa = 3.35NFNSFNTDD46 pKa = 1.49IWNIDD51 pKa = 3.6EE52 pKa = 5.48GDD54 pKa = 3.44GCAQGLCGWGNQEE67 pKa = 4.2LQWYY71 pKa = 8.36AQDD74 pKa = 3.28NVYY77 pKa = 10.32IQDD80 pKa = 3.5IAGEE84 pKa = 4.22PGNKK88 pKa = 9.8GLVLEE93 pKa = 5.12ARR95 pKa = 11.84NQATGGKK102 pKa = 9.33AFTSGKK108 pKa = 8.98IQSSNKK114 pKa = 9.67LAIKK118 pKa = 10.41YY119 pKa = 10.3GMIEE123 pKa = 4.22MRR125 pKa = 11.84TKK127 pKa = 10.11TPNVDD132 pKa = 3.05TGLWPALWMLGTSTSSWPAKK152 pKa = 10.55GEE154 pKa = 3.72IDD156 pKa = 3.56IMEE159 pKa = 4.65MGQSAAQRR167 pKa = 11.84ADD169 pKa = 2.91AGFPGAPLNNYY180 pKa = 8.87VGSNLIFYY188 pKa = 10.75ADD190 pKa = 3.98AACSDD195 pKa = 4.32GNPSCAASTAWQTDD209 pKa = 3.33NAHH212 pKa = 6.6LSSSSLANRR221 pKa = 11.84FVTYY225 pKa = 10.45RR226 pKa = 11.84LYY228 pKa = 8.99WTEE231 pKa = 3.69SQIRR235 pKa = 11.84FAIVDD240 pKa = 3.49GGVEE244 pKa = 4.22YY245 pKa = 10.82EE246 pKa = 4.7MYY248 pKa = 10.26SQPFTISEE256 pKa = 4.2EE257 pKa = 4.36SNEE260 pKa = 4.09FQQPFYY266 pKa = 11.42LLMDD270 pKa = 4.7LAVGGTFTDD279 pKa = 3.53AQTNGQVTAPLPAKK293 pKa = 9.83MVVDD297 pKa = 3.68YY298 pKa = 11.41VRR300 pKa = 11.84IYY302 pKa = 10.69EE303 pKa = 4.35LDD305 pKa = 3.53GQGEE309 pKa = 4.3IFLGNTVPEE318 pKa = 4.17EE319 pKa = 3.99TGTFGVFTDD328 pKa = 3.91NTPTNNKK335 pKa = 9.73LEE337 pKa = 4.74PGVDD341 pKa = 2.94ADD343 pKa = 3.66IYY345 pKa = 10.96VWNQTSITDD354 pKa = 3.81GTTPPYY360 pKa = 10.85EE361 pKa = 4.71GDD363 pKa = 4.67NVLAWQYY370 pKa = 11.63SPSQWFGGGFQSRR383 pKa = 11.84QPRR386 pKa = 11.84DD387 pKa = 3.25LSNFADD393 pKa = 3.73GDD395 pKa = 3.91VHH397 pKa = 7.15FKK399 pKa = 10.65IKK401 pKa = 10.37IPADD405 pKa = 3.23VSFKK409 pKa = 11.22VGIADD414 pKa = 4.3TYY416 pKa = 10.92TNEE419 pKa = 3.53NWIEE423 pKa = 4.05FPANTTKK430 pKa = 10.93YY431 pKa = 9.91GLVRR435 pKa = 11.84NGDD438 pKa = 3.16WAEE441 pKa = 3.82AVIPVSDD448 pKa = 3.76LAGPLVALQSISNPFNIASVDD469 pKa = 3.84GQIPTYY475 pKa = 9.98AFEE478 pKa = 4.17MALDD482 pKa = 4.81DD483 pKa = 4.72IVWTGGGGPVVQDD496 pKa = 3.64SDD498 pKa = 4.09GDD500 pKa = 4.33GINDD504 pKa = 4.87NDD506 pKa = 4.31DD507 pKa = 3.56QCPTTAGDD515 pKa = 3.84AANNGCPASSNAIDD529 pKa = 3.94LTDD532 pKa = 3.49VPGTFTAQHH541 pKa = 6.39NDD543 pKa = 2.85SPANEE548 pKa = 4.18GAEE551 pKa = 4.02NAFDD555 pKa = 6.02DD556 pKa = 4.52DD557 pKa = 4.19TTTKK561 pKa = 10.84YY562 pKa = 9.94LTFHH566 pKa = 6.69ASAWLQYY573 pKa = 8.61QTADD577 pKa = 3.25FAYY580 pKa = 8.35PASGYY585 pKa = 10.41RR586 pKa = 11.84ITSANDD592 pKa = 3.06APEE595 pKa = 4.88RR596 pKa = 11.84DD597 pKa = 3.51PMSWNFQGSSDD608 pKa = 3.56GVTWTTLDD616 pKa = 3.37SRR618 pKa = 11.84SNEE621 pKa = 3.93DD622 pKa = 2.94FSSRR626 pKa = 11.84FQTRR630 pKa = 11.84EE631 pKa = 3.45FSFSNTTAWQFYY643 pKa = 9.35RR644 pKa = 11.84INMMNNSGGMTQVAEE659 pKa = 4.15FSILGSDD666 pKa = 3.47NPVPVNQNSDD676 pKa = 3.17SDD678 pKa = 4.17GDD680 pKa = 4.23GVADD684 pKa = 4.9SADD687 pKa = 3.66NCPDD691 pKa = 3.54TPAGTQVDD699 pKa = 4.42GNGCAISDD707 pKa = 3.72GATGVEE713 pKa = 4.24QVSADD718 pKa = 3.61SIVFFVNTADD728 pKa = 3.52WADD731 pKa = 3.21VHH733 pKa = 5.46YY734 pKa = 9.45TINGQNQQNLRR745 pKa = 11.84MNIEE749 pKa = 3.9AGRR752 pKa = 11.84NEE754 pKa = 3.79TRR756 pKa = 11.84VNGLSAGDD764 pKa = 4.05TITYY768 pKa = 7.87WFTYY772 pKa = 8.25LQPNGAVTDD781 pKa = 4.23TSPQTYY787 pKa = 10.27SVVTTSAGDD796 pKa = 3.59ADD798 pKa = 4.11GDD800 pKa = 4.31GVNDD804 pKa = 5.76GVDD807 pKa = 3.2QCANTPAGASVDD819 pKa = 3.66EE820 pKa = 4.93SGCEE824 pKa = 3.92TLVTTSVVVEE834 pKa = 4.07AEE836 pKa = 4.61NYY838 pKa = 10.49SNWNDD843 pKa = 2.92SDD845 pKa = 3.58AGNNGGAYY853 pKa = 10.4RR854 pKa = 11.84NDD856 pKa = 3.75DD857 pKa = 3.43VDD859 pKa = 4.4IEE861 pKa = 4.35PTTDD865 pKa = 2.76INSGYY870 pKa = 9.7NVGWTASGEE879 pKa = 4.06WLEE882 pKa = 4.16YY883 pKa = 10.86AVTLGAGTYY892 pKa = 8.72QVSSRR897 pKa = 11.84VASNTGAGGYY907 pKa = 7.62TVALNGAEE915 pKa = 3.92FATDD919 pKa = 3.28NVEE922 pKa = 4.77ATGGWQSFVTHH933 pKa = 6.54SLGQVTITEE942 pKa = 4.85GGSQTLRR949 pKa = 11.84INFTGADD956 pKa = 3.62VNFNWLKK963 pKa = 10.98FEE965 pKa = 4.82LLGNN969 pKa = 4.2
MM1 pKa = 7.73RR2 pKa = 11.84KK3 pKa = 8.14ITLSSRR9 pKa = 11.84QLSKK13 pKa = 10.66IALASALAFCSSQSHH28 pKa = 5.84AAVGNLIWEE37 pKa = 4.7DD38 pKa = 3.35NFNSFNTDD46 pKa = 1.49IWNIDD51 pKa = 3.6EE52 pKa = 5.48GDD54 pKa = 3.44GCAQGLCGWGNQEE67 pKa = 4.2LQWYY71 pKa = 8.36AQDD74 pKa = 3.28NVYY77 pKa = 10.32IQDD80 pKa = 3.5IAGEE84 pKa = 4.22PGNKK88 pKa = 9.8GLVLEE93 pKa = 5.12ARR95 pKa = 11.84NQATGGKK102 pKa = 9.33AFTSGKK108 pKa = 8.98IQSSNKK114 pKa = 9.67LAIKK118 pKa = 10.41YY119 pKa = 10.3GMIEE123 pKa = 4.22MRR125 pKa = 11.84TKK127 pKa = 10.11TPNVDD132 pKa = 3.05TGLWPALWMLGTSTSSWPAKK152 pKa = 10.55GEE154 pKa = 3.72IDD156 pKa = 3.56IMEE159 pKa = 4.65MGQSAAQRR167 pKa = 11.84ADD169 pKa = 2.91AGFPGAPLNNYY180 pKa = 8.87VGSNLIFYY188 pKa = 10.75ADD190 pKa = 3.98AACSDD195 pKa = 4.32GNPSCAASTAWQTDD209 pKa = 3.33NAHH212 pKa = 6.6LSSSSLANRR221 pKa = 11.84FVTYY225 pKa = 10.45RR226 pKa = 11.84LYY228 pKa = 8.99WTEE231 pKa = 3.69SQIRR235 pKa = 11.84FAIVDD240 pKa = 3.49GGVEE244 pKa = 4.22YY245 pKa = 10.82EE246 pKa = 4.7MYY248 pKa = 10.26SQPFTISEE256 pKa = 4.2EE257 pKa = 4.36SNEE260 pKa = 4.09FQQPFYY266 pKa = 11.42LLMDD270 pKa = 4.7LAVGGTFTDD279 pKa = 3.53AQTNGQVTAPLPAKK293 pKa = 9.83MVVDD297 pKa = 3.68YY298 pKa = 11.41VRR300 pKa = 11.84IYY302 pKa = 10.69EE303 pKa = 4.35LDD305 pKa = 3.53GQGEE309 pKa = 4.3IFLGNTVPEE318 pKa = 4.17EE319 pKa = 3.99TGTFGVFTDD328 pKa = 3.91NTPTNNKK335 pKa = 9.73LEE337 pKa = 4.74PGVDD341 pKa = 2.94ADD343 pKa = 3.66IYY345 pKa = 10.96VWNQTSITDD354 pKa = 3.81GTTPPYY360 pKa = 10.85EE361 pKa = 4.71GDD363 pKa = 4.67NVLAWQYY370 pKa = 11.63SPSQWFGGGFQSRR383 pKa = 11.84QPRR386 pKa = 11.84DD387 pKa = 3.25LSNFADD393 pKa = 3.73GDD395 pKa = 3.91VHH397 pKa = 7.15FKK399 pKa = 10.65IKK401 pKa = 10.37IPADD405 pKa = 3.23VSFKK409 pKa = 11.22VGIADD414 pKa = 4.3TYY416 pKa = 10.92TNEE419 pKa = 3.53NWIEE423 pKa = 4.05FPANTTKK430 pKa = 10.93YY431 pKa = 9.91GLVRR435 pKa = 11.84NGDD438 pKa = 3.16WAEE441 pKa = 3.82AVIPVSDD448 pKa = 3.76LAGPLVALQSISNPFNIASVDD469 pKa = 3.84GQIPTYY475 pKa = 9.98AFEE478 pKa = 4.17MALDD482 pKa = 4.81DD483 pKa = 4.72IVWTGGGGPVVQDD496 pKa = 3.64SDD498 pKa = 4.09GDD500 pKa = 4.33GINDD504 pKa = 4.87NDD506 pKa = 4.31DD507 pKa = 3.56QCPTTAGDD515 pKa = 3.84AANNGCPASSNAIDD529 pKa = 3.94LTDD532 pKa = 3.49VPGTFTAQHH541 pKa = 6.39NDD543 pKa = 2.85SPANEE548 pKa = 4.18GAEE551 pKa = 4.02NAFDD555 pKa = 6.02DD556 pKa = 4.52DD557 pKa = 4.19TTTKK561 pKa = 10.84YY562 pKa = 9.94LTFHH566 pKa = 6.69ASAWLQYY573 pKa = 8.61QTADD577 pKa = 3.25FAYY580 pKa = 8.35PASGYY585 pKa = 10.41RR586 pKa = 11.84ITSANDD592 pKa = 3.06APEE595 pKa = 4.88RR596 pKa = 11.84DD597 pKa = 3.51PMSWNFQGSSDD608 pKa = 3.56GVTWTTLDD616 pKa = 3.37SRR618 pKa = 11.84SNEE621 pKa = 3.93DD622 pKa = 2.94FSSRR626 pKa = 11.84FQTRR630 pKa = 11.84EE631 pKa = 3.45FSFSNTTAWQFYY643 pKa = 9.35RR644 pKa = 11.84INMMNNSGGMTQVAEE659 pKa = 4.15FSILGSDD666 pKa = 3.47NPVPVNQNSDD676 pKa = 3.17SDD678 pKa = 4.17GDD680 pKa = 4.23GVADD684 pKa = 4.9SADD687 pKa = 3.66NCPDD691 pKa = 3.54TPAGTQVDD699 pKa = 4.42GNGCAISDD707 pKa = 3.72GATGVEE713 pKa = 4.24QVSADD718 pKa = 3.61SIVFFVNTADD728 pKa = 3.52WADD731 pKa = 3.21VHH733 pKa = 5.46YY734 pKa = 9.45TINGQNQQNLRR745 pKa = 11.84MNIEE749 pKa = 3.9AGRR752 pKa = 11.84NEE754 pKa = 3.79TRR756 pKa = 11.84VNGLSAGDD764 pKa = 4.05TITYY768 pKa = 7.87WFTYY772 pKa = 8.25LQPNGAVTDD781 pKa = 4.23TSPQTYY787 pKa = 10.27SVVTTSAGDD796 pKa = 3.59ADD798 pKa = 4.11GDD800 pKa = 4.31GVNDD804 pKa = 5.76GVDD807 pKa = 3.2QCANTPAGASVDD819 pKa = 3.66EE820 pKa = 4.93SGCEE824 pKa = 3.92TLVTTSVVVEE834 pKa = 4.07AEE836 pKa = 4.61NYY838 pKa = 10.49SNWNDD843 pKa = 2.92SDD845 pKa = 3.58AGNNGGAYY853 pKa = 10.4RR854 pKa = 11.84NDD856 pKa = 3.75DD857 pKa = 3.43VDD859 pKa = 4.4IEE861 pKa = 4.35PTTDD865 pKa = 2.76INSGYY870 pKa = 9.7NVGWTASGEE879 pKa = 4.06WLEE882 pKa = 4.16YY883 pKa = 10.86AVTLGAGTYY892 pKa = 8.72QVSSRR897 pKa = 11.84VASNTGAGGYY907 pKa = 7.62TVALNGAEE915 pKa = 3.92FATDD919 pKa = 3.28NVEE922 pKa = 4.77ATGGWQSFVTHH933 pKa = 6.54SLGQVTITEE942 pKa = 4.85GGSQTLRR949 pKa = 11.84INFTGADD956 pKa = 3.62VNFNWLKK963 pKa = 10.98FEE965 pKa = 4.82LLGNN969 pKa = 4.2
Molecular weight: 104.03 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6M4MDS1|A0A6M4MDS1_9ALTE Uncharacterized protein OS=Alteromonas pelagimontana OX=1858656 GN=CA267_010895 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 10.14RR12 pKa = 11.84KK13 pKa = 9.13RR14 pKa = 11.84SHH16 pKa = 6.17GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37NGRR28 pKa = 11.84KK29 pKa = 9.16VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.28GRR39 pKa = 11.84ARR41 pKa = 11.84LCAA44 pKa = 3.92
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 10.14RR12 pKa = 11.84KK13 pKa = 9.13RR14 pKa = 11.84SHH16 pKa = 6.17GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37NGRR28 pKa = 11.84KK29 pKa = 9.16VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.28GRR39 pKa = 11.84ARR41 pKa = 11.84LCAA44 pKa = 3.92
Molecular weight: 5.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1214117 |
22 |
2045 |
335.7 |
37.26 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.19 ± 0.041 | 0.977 ± 0.015 |
5.676 ± 0.035 | 6.187 ± 0.036 |
4.188 ± 0.028 | 6.836 ± 0.044 |
2.249 ± 0.02 | 6.047 ± 0.028 |
4.98 ± 0.033 | 9.996 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.517 ± 0.02 | 4.243 ± 0.032 |
4.092 ± 0.024 | 4.48 ± 0.034 |
4.868 ± 0.031 | 6.53 ± 0.03 |
5.485 ± 0.03 | 7.011 ± 0.03 |
1.315 ± 0.018 | 3.132 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
