
Alcanivorax dieselolei (strain DSM 16502 / CGMCC 1.3690 / B-5)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Oceanospirillales; Alcanivoracaceae; Alcanivorax; Alcanivorax dieselolei
Average proteome isoelectric point is 6.3
Get precalculated fractions of proteins
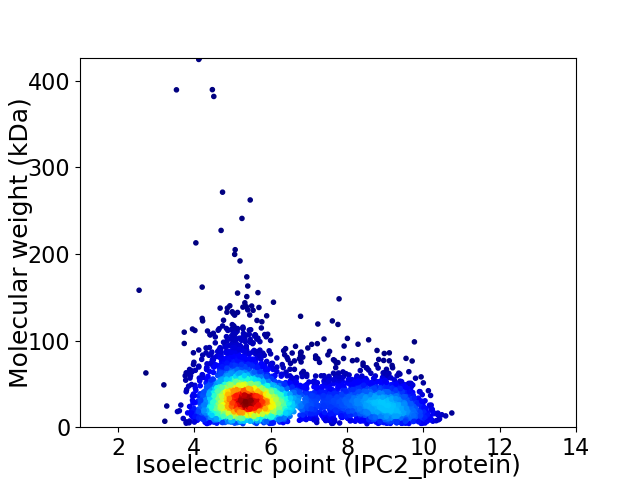
Virtual 2D-PAGE plot for 4388 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|K0CCK1|K0CCK1_ALCDB Erythronate-4-phosphate dehydrogenase OS=Alcanivorax dieselolei (strain DSM 16502 / CGMCC 1.3690 / B-5) OX=930169 GN=pdxB PE=3 SV=1
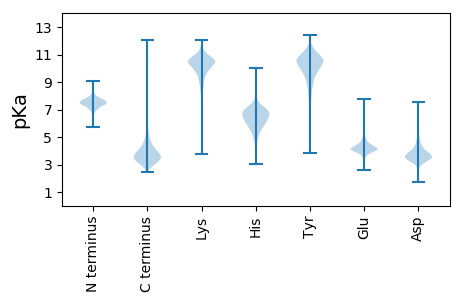
MM1 pKa = 7.36IKK3 pKa = 10.17KK4 pKa = 9.77AALAAAIASAPFAASATDD22 pKa = 3.79LVGGGATLPAVAYY35 pKa = 10.07VGEE38 pKa = 5.16DD39 pKa = 3.39FTLTDD44 pKa = 3.54PQARR48 pKa = 11.84LSTNAGLNPGSGFLDD63 pKa = 3.71APLGIDD69 pKa = 3.44SIFDD73 pKa = 3.31VFEE76 pKa = 4.18QNNLSHH82 pKa = 6.33TASYY86 pKa = 10.14CQTGSSTGKK95 pKa = 10.08RR96 pKa = 11.84VLLGASGWSASGNCRR111 pKa = 11.84DD112 pKa = 4.2YY113 pKa = 11.6SDD115 pKa = 4.11SPEE118 pKa = 4.28GFSGLDD124 pKa = 3.5ALPDD128 pKa = 3.96FAGSDD133 pKa = 3.79APLTADD139 pKa = 5.79DD140 pKa = 5.16IVTFQNGNEE149 pKa = 3.98AARR152 pKa = 11.84TNLVQVPALGAFLGLPINIQDD173 pKa = 3.66GSGNPITNVDD183 pKa = 3.66LSTAEE188 pKa = 3.7VCAIFDD194 pKa = 3.69GSATVWSDD202 pKa = 2.93VGVPSSQQIKK212 pKa = 9.61VVHH215 pKa = 6.51RR216 pKa = 11.84SDD218 pKa = 3.96GSGTTFAFSQYY229 pKa = 10.6LAANCNSGLAPTFTTQEE246 pKa = 4.35LYY248 pKa = 11.13SDD250 pKa = 5.32AISLTVYY257 pKa = 10.32GGRR260 pKa = 11.84DD261 pKa = 3.22VGGSGNGGVIDD272 pKa = 4.92AVTGTPGAIGYY283 pKa = 10.9ANFSNVAVEE292 pKa = 4.26PGLAYY297 pKa = 10.53ALVEE301 pKa = 4.2GHH303 pKa = 7.11DD304 pKa = 3.92PANGGSINVAASDD317 pKa = 3.55ILIDD321 pKa = 5.43KK322 pKa = 10.54VLGDD326 pKa = 3.89NDD328 pKa = 4.44SVTGLPKK335 pKa = 10.53PEE337 pKa = 4.94DD338 pKa = 3.81VDD340 pKa = 4.11SSLAQEE346 pKa = 4.55CLAVVDD352 pKa = 4.47PAVQLEE358 pKa = 4.1QVYY361 pKa = 9.7PIVAFTNLLTYY372 pKa = 9.56TEE374 pKa = 4.59NNNDD378 pKa = 3.73PAATQALFTEE388 pKa = 5.15VTHH391 pKa = 6.3GTRR394 pKa = 11.84DD395 pKa = 3.49QNGEE399 pKa = 4.06PVYY402 pKa = 10.24DD403 pKa = 4.09LPVGYY408 pKa = 10.73ARR410 pKa = 11.84VASTAVSNIISTCINN425 pKa = 3.18
MM1 pKa = 7.36IKK3 pKa = 10.17KK4 pKa = 9.77AALAAAIASAPFAASATDD22 pKa = 3.79LVGGGATLPAVAYY35 pKa = 10.07VGEE38 pKa = 5.16DD39 pKa = 3.39FTLTDD44 pKa = 3.54PQARR48 pKa = 11.84LSTNAGLNPGSGFLDD63 pKa = 3.71APLGIDD69 pKa = 3.44SIFDD73 pKa = 3.31VFEE76 pKa = 4.18QNNLSHH82 pKa = 6.33TASYY86 pKa = 10.14CQTGSSTGKK95 pKa = 10.08RR96 pKa = 11.84VLLGASGWSASGNCRR111 pKa = 11.84DD112 pKa = 4.2YY113 pKa = 11.6SDD115 pKa = 4.11SPEE118 pKa = 4.28GFSGLDD124 pKa = 3.5ALPDD128 pKa = 3.96FAGSDD133 pKa = 3.79APLTADD139 pKa = 5.79DD140 pKa = 5.16IVTFQNGNEE149 pKa = 3.98AARR152 pKa = 11.84TNLVQVPALGAFLGLPINIQDD173 pKa = 3.66GSGNPITNVDD183 pKa = 3.66LSTAEE188 pKa = 3.7VCAIFDD194 pKa = 3.69GSATVWSDD202 pKa = 2.93VGVPSSQQIKK212 pKa = 9.61VVHH215 pKa = 6.51RR216 pKa = 11.84SDD218 pKa = 3.96GSGTTFAFSQYY229 pKa = 10.6LAANCNSGLAPTFTTQEE246 pKa = 4.35LYY248 pKa = 11.13SDD250 pKa = 5.32AISLTVYY257 pKa = 10.32GGRR260 pKa = 11.84DD261 pKa = 3.22VGGSGNGGVIDD272 pKa = 4.92AVTGTPGAIGYY283 pKa = 10.9ANFSNVAVEE292 pKa = 4.26PGLAYY297 pKa = 10.53ALVEE301 pKa = 4.2GHH303 pKa = 7.11DD304 pKa = 3.92PANGGSINVAASDD317 pKa = 3.55ILIDD321 pKa = 5.43KK322 pKa = 10.54VLGDD326 pKa = 3.89NDD328 pKa = 4.44SVTGLPKK335 pKa = 10.53PEE337 pKa = 4.94DD338 pKa = 3.81VDD340 pKa = 4.11SSLAQEE346 pKa = 4.55CLAVVDD352 pKa = 4.47PAVQLEE358 pKa = 4.1QVYY361 pKa = 9.7PIVAFTNLLTYY372 pKa = 9.56TEE374 pKa = 4.59NNNDD378 pKa = 3.73PAATQALFTEE388 pKa = 5.15VTHH391 pKa = 6.3GTRR394 pKa = 11.84DD395 pKa = 3.49QNGEE399 pKa = 4.06PVYY402 pKa = 10.24DD403 pKa = 4.09LPVGYY408 pKa = 10.73ARR410 pKa = 11.84VASTAVSNIISTCINN425 pKa = 3.18
Molecular weight: 43.38 kDa
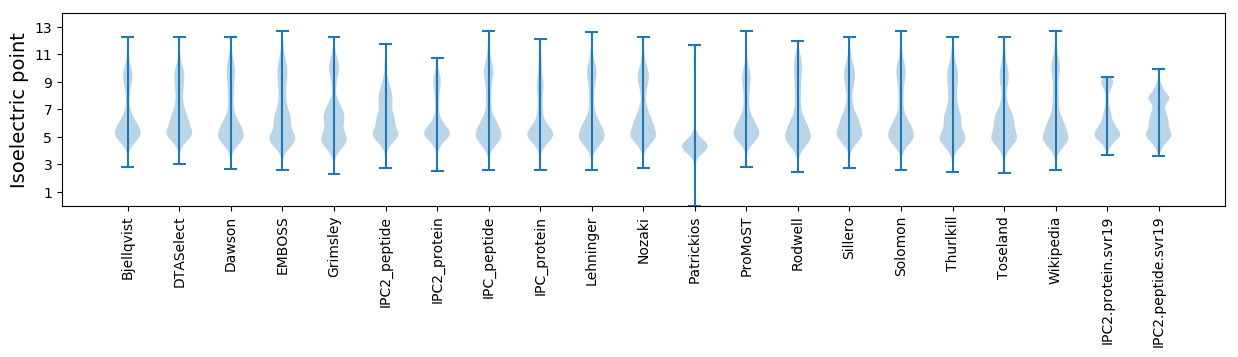
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|K0CDP3|K0CDP3_ALCDB Molybdate ABC transporter periplasmic molybdate-binding protein OS=Alcanivorax dieselolei (strain DSM 16502 / CGMCC 1.3690 / B-5) OX=930169 GN=B5T_02429 PE=3 SV=1
MM1 pKa = 7.77ALPSSTKK8 pKa = 10.1SDD10 pKa = 3.6LLLVCVTLLAAVSWMFSKK28 pKa = 10.62EE29 pKa = 3.99AVLLMPPLLFMCLRR43 pKa = 11.84FLLAGLILALVGQRR57 pKa = 11.84QLRR60 pKa = 11.84RR61 pKa = 11.84MNRR64 pKa = 11.84DD65 pKa = 3.08KK66 pKa = 10.75YY67 pKa = 9.19WRR69 pKa = 11.84SIQVGMVFGAAMSFWIMGLHH89 pKa = 6.02YY90 pKa = 10.07GNHH93 pKa = 5.6VGEE96 pKa = 4.67GAFITSLGVVLVPVMARR113 pKa = 11.84LVFKK117 pKa = 8.46EE118 pKa = 4.24HH119 pKa = 7.09PPLSTWLALPVAVCGLALLSLDD141 pKa = 4.02GGFQGEE147 pKa = 4.4LGQLFFLASAMIFALFFTLNTRR169 pKa = 11.84AANNATITATGGRR182 pKa = 11.84TTTRR186 pKa = 11.84EE187 pKa = 3.87RR188 pKa = 11.84VPALALTSVVLVTVGLVTGTASLLLEE214 pKa = 4.2PWSPTFHH221 pKa = 7.36SGEE224 pKa = 4.21LGLVIAAWVIASATLGTAARR244 pKa = 11.84FFLQTYY250 pKa = 7.94AQSLSMHH257 pKa = 5.21SHH259 pKa = 5.87GVVIMILEE267 pKa = 4.25PMWTALLAALWFGEE281 pKa = 4.32TMNATQLGGCAMIFLALLVNRR302 pKa = 11.84WSAIRR307 pKa = 11.84RR308 pKa = 11.84VLKK311 pKa = 10.25RR312 pKa = 11.84WVDD315 pKa = 3.25QRR317 pKa = 11.84RR318 pKa = 11.84AARR321 pKa = 11.84AA322 pKa = 3.18
MM1 pKa = 7.77ALPSSTKK8 pKa = 10.1SDD10 pKa = 3.6LLLVCVTLLAAVSWMFSKK28 pKa = 10.62EE29 pKa = 3.99AVLLMPPLLFMCLRR43 pKa = 11.84FLLAGLILALVGQRR57 pKa = 11.84QLRR60 pKa = 11.84RR61 pKa = 11.84MNRR64 pKa = 11.84DD65 pKa = 3.08KK66 pKa = 10.75YY67 pKa = 9.19WRR69 pKa = 11.84SIQVGMVFGAAMSFWIMGLHH89 pKa = 6.02YY90 pKa = 10.07GNHH93 pKa = 5.6VGEE96 pKa = 4.67GAFITSLGVVLVPVMARR113 pKa = 11.84LVFKK117 pKa = 8.46EE118 pKa = 4.24HH119 pKa = 7.09PPLSTWLALPVAVCGLALLSLDD141 pKa = 4.02GGFQGEE147 pKa = 4.4LGQLFFLASAMIFALFFTLNTRR169 pKa = 11.84AANNATITATGGRR182 pKa = 11.84TTTRR186 pKa = 11.84EE187 pKa = 3.87RR188 pKa = 11.84VPALALTSVVLVTVGLVTGTASLLLEE214 pKa = 4.2PWSPTFHH221 pKa = 7.36SGEE224 pKa = 4.21LGLVIAAWVIASATLGTAARR244 pKa = 11.84FFLQTYY250 pKa = 7.94AQSLSMHH257 pKa = 5.21SHH259 pKa = 5.87GVVIMILEE267 pKa = 4.25PMWTALLAALWFGEE281 pKa = 4.32TMNATQLGGCAMIFLALLVNRR302 pKa = 11.84WSAIRR307 pKa = 11.84RR308 pKa = 11.84VLKK311 pKa = 10.25RR312 pKa = 11.84WVDD315 pKa = 3.25QRR317 pKa = 11.84RR318 pKa = 11.84AARR321 pKa = 11.84AA322 pKa = 3.18
Molecular weight: 35.05 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1455496 |
37 |
4089 |
331.7 |
36.45 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.699 ± 0.049 | 0.918 ± 0.013 |
5.987 ± 0.036 | 6.053 ± 0.036 |
3.496 ± 0.024 | 8.457 ± 0.051 |
2.3 ± 0.021 | 4.375 ± 0.027 |
2.859 ± 0.033 | 11.229 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.42 ± 0.021 | 2.858 ± 0.024 |
5.054 ± 0.03 | 4.037 ± 0.026 |
7.332 ± 0.043 | 5.374 ± 0.03 |
5.072 ± 0.031 | 7.379 ± 0.034 |
1.522 ± 0.016 | 2.581 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
