
Influenza C virus (strain C/Ann Arbor/1/1950)
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Polyploviricotina; Insthoviricetes; Articulavirales; Orthomyxoviridae; Gammainfluenzavirus; Influenza C virus
Average proteome isoelectric point is 7.34
Get precalculated fractions of proteins
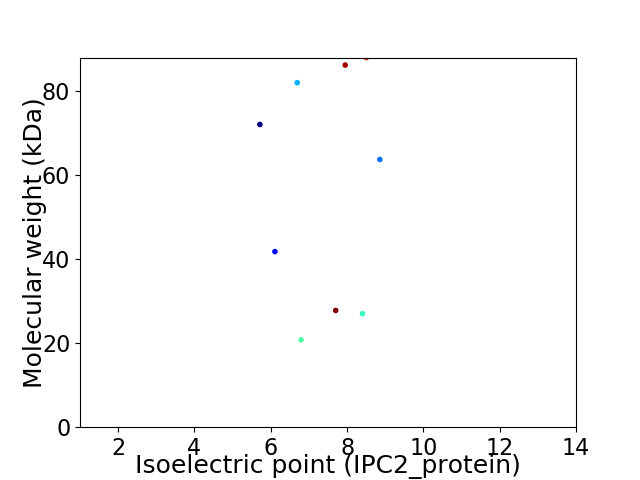
Virtual 2D-PAGE plot for 9 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
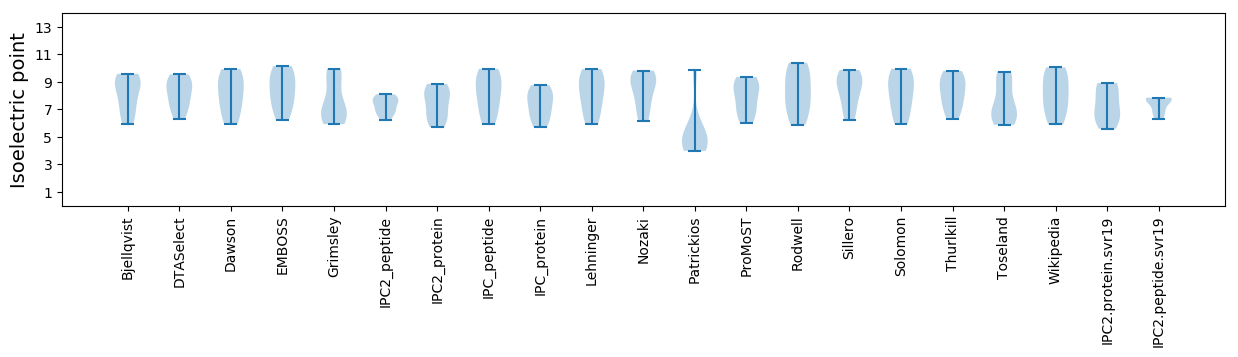
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q01639|NS1_INCAA Non-structural protein 1 OS=Influenza C virus (strain C/Ann Arbor/1/1950) OX=11553 GN=NS PE=2 SV=2
MM1 pKa = 7.28FFSLLLMLGLTEE13 pKa = 4.17AEE15 pKa = 4.5KK16 pKa = 10.71IKK18 pKa = 10.49ICLQKK23 pKa = 10.46QVNSSFSLHH32 pKa = 6.18NGFGGNLYY40 pKa = 9.14ATEE43 pKa = 4.17EE44 pKa = 3.89KK45 pKa = 10.88RR46 pKa = 11.84MFEE49 pKa = 4.06LVKK52 pKa = 10.01PKK54 pKa = 10.77AGASVLNQSTWIGFGDD70 pKa = 3.97SRR72 pKa = 11.84TDD74 pKa = 3.11KK75 pKa = 11.14SNSAFPRR82 pKa = 11.84SADD85 pKa = 3.48VSAKK89 pKa = 9.58TADD92 pKa = 3.48KK93 pKa = 10.49FRR95 pKa = 11.84SLSGGSLMLSMFGPPGKK112 pKa = 9.91VDD114 pKa = 3.66YY115 pKa = 10.63LYY117 pKa = 10.65QGCGKK122 pKa = 9.87HH123 pKa = 5.19KK124 pKa = 10.57VFYY127 pKa = 10.67EE128 pKa = 4.31GVNWSPHH135 pKa = 4.95AAINCYY141 pKa = 9.54RR142 pKa = 11.84KK143 pKa = 9.49NWTDD147 pKa = 2.75IKK149 pKa = 11.35LNFQKK154 pKa = 10.74NIYY157 pKa = 8.76EE158 pKa = 4.29LASQSHH164 pKa = 6.17CMSLVNALDD173 pKa = 3.42KK174 pKa = 10.47TIPLQATAGVAKK186 pKa = 10.38NCNNSFLKK194 pKa = 10.92NPALYY199 pKa = 8.36TQEE202 pKa = 4.12VNPSVEE208 pKa = 4.03KK209 pKa = 10.76CGKK212 pKa = 9.53EE213 pKa = 3.49NLAFFTLPTQFGTYY227 pKa = 7.66EE228 pKa = 4.29CKK230 pKa = 10.21LHH232 pKa = 6.73LVASCYY238 pKa = 10.19FIYY241 pKa = 10.31DD242 pKa = 3.38SKK244 pKa = 11.26EE245 pKa = 3.89VYY247 pKa = 9.91NKK249 pKa = 10.22RR250 pKa = 11.84GCDD253 pKa = 2.93NYY255 pKa = 10.72FQVIYY260 pKa = 10.21DD261 pKa = 3.56SSGKK265 pKa = 10.06VVGGLDD271 pKa = 3.6NRR273 pKa = 11.84VSPYY277 pKa = 9.64TGNSGDD283 pKa = 4.18TPTMQCDD290 pKa = 3.55MLQLKK295 pKa = 9.31PGRR298 pKa = 11.84YY299 pKa = 8.55SVRR302 pKa = 11.84SSPRR306 pKa = 11.84FLLMPEE312 pKa = 4.1RR313 pKa = 11.84SYY315 pKa = 11.73CFDD318 pKa = 3.5MKK320 pKa = 10.99EE321 pKa = 4.16KK322 pKa = 11.04GPVTAVQSIWGKK334 pKa = 10.35GRR336 pKa = 11.84EE337 pKa = 4.07SDD339 pKa = 3.62HH340 pKa = 7.93AVDD343 pKa = 4.18QACLSTPGCMLIQKK357 pKa = 7.37QKK359 pKa = 10.35PYY361 pKa = 10.24IGEE364 pKa = 4.33ADD366 pKa = 3.72DD367 pKa = 3.83HH368 pKa = 7.68HH369 pKa = 7.57GDD371 pKa = 3.26QEE373 pKa = 4.19MRR375 pKa = 11.84EE376 pKa = 4.26LLSGLDD382 pKa = 3.52YY383 pKa = 10.79EE384 pKa = 5.02ARR386 pKa = 11.84CISQSGWVNEE396 pKa = 4.0TSPFTEE402 pKa = 5.12EE403 pKa = 3.88YY404 pKa = 10.82LLPPKK409 pKa = 10.63FGRR412 pKa = 11.84CPLAAKK418 pKa = 10.04EE419 pKa = 3.87EE420 pKa = 4.69SIPKK424 pKa = 9.95IPDD427 pKa = 3.22GLLIPTSGTDD437 pKa = 3.31TTVTKK442 pKa = 10.09PKK444 pKa = 10.63SRR446 pKa = 11.84IFGIDD451 pKa = 3.38DD452 pKa = 5.09LIIGLLFVAIVEE464 pKa = 4.27AGIGGYY470 pKa = 10.53LLGSRR475 pKa = 11.84KK476 pKa = 9.78VSGGGVTKK484 pKa = 10.59EE485 pKa = 3.9SAEE488 pKa = 4.1KK489 pKa = 10.63GFEE492 pKa = 4.3KK493 pKa = 10.09IGNDD497 pKa = 3.11IQILRR502 pKa = 11.84SSTNIAIEE510 pKa = 4.02KK511 pKa = 9.93LNDD514 pKa = 4.38RR515 pKa = 11.84ISHH518 pKa = 6.87DD519 pKa = 3.58EE520 pKa = 3.67QAIRR524 pKa = 11.84DD525 pKa = 3.81LTLEE529 pKa = 4.02IEE531 pKa = 4.48NARR534 pKa = 11.84SEE536 pKa = 4.27ALLGEE541 pKa = 4.29LGIIRR546 pKa = 11.84ALLVGNISIGLQEE559 pKa = 4.78SLWEE563 pKa = 3.97LASEE567 pKa = 4.14ITNRR571 pKa = 11.84AGDD574 pKa = 3.74LAVEE578 pKa = 4.25VSPGCWVIDD587 pKa = 4.49NNICDD592 pKa = 3.64QSCQNFIFKK601 pKa = 10.49FNEE604 pKa = 3.92TAPVPTIPPLDD615 pKa = 3.72TKK617 pKa = 10.71IDD619 pKa = 3.76LQSDD623 pKa = 4.09PFYY626 pKa = 10.2WGSSLGLAITAAISLAALVISGIAICRR653 pKa = 11.84TKK655 pKa = 11.06
MM1 pKa = 7.28FFSLLLMLGLTEE13 pKa = 4.17AEE15 pKa = 4.5KK16 pKa = 10.71IKK18 pKa = 10.49ICLQKK23 pKa = 10.46QVNSSFSLHH32 pKa = 6.18NGFGGNLYY40 pKa = 9.14ATEE43 pKa = 4.17EE44 pKa = 3.89KK45 pKa = 10.88RR46 pKa = 11.84MFEE49 pKa = 4.06LVKK52 pKa = 10.01PKK54 pKa = 10.77AGASVLNQSTWIGFGDD70 pKa = 3.97SRR72 pKa = 11.84TDD74 pKa = 3.11KK75 pKa = 11.14SNSAFPRR82 pKa = 11.84SADD85 pKa = 3.48VSAKK89 pKa = 9.58TADD92 pKa = 3.48KK93 pKa = 10.49FRR95 pKa = 11.84SLSGGSLMLSMFGPPGKK112 pKa = 9.91VDD114 pKa = 3.66YY115 pKa = 10.63LYY117 pKa = 10.65QGCGKK122 pKa = 9.87HH123 pKa = 5.19KK124 pKa = 10.57VFYY127 pKa = 10.67EE128 pKa = 4.31GVNWSPHH135 pKa = 4.95AAINCYY141 pKa = 9.54RR142 pKa = 11.84KK143 pKa = 9.49NWTDD147 pKa = 2.75IKK149 pKa = 11.35LNFQKK154 pKa = 10.74NIYY157 pKa = 8.76EE158 pKa = 4.29LASQSHH164 pKa = 6.17CMSLVNALDD173 pKa = 3.42KK174 pKa = 10.47TIPLQATAGVAKK186 pKa = 10.38NCNNSFLKK194 pKa = 10.92NPALYY199 pKa = 8.36TQEE202 pKa = 4.12VNPSVEE208 pKa = 4.03KK209 pKa = 10.76CGKK212 pKa = 9.53EE213 pKa = 3.49NLAFFTLPTQFGTYY227 pKa = 7.66EE228 pKa = 4.29CKK230 pKa = 10.21LHH232 pKa = 6.73LVASCYY238 pKa = 10.19FIYY241 pKa = 10.31DD242 pKa = 3.38SKK244 pKa = 11.26EE245 pKa = 3.89VYY247 pKa = 9.91NKK249 pKa = 10.22RR250 pKa = 11.84GCDD253 pKa = 2.93NYY255 pKa = 10.72FQVIYY260 pKa = 10.21DD261 pKa = 3.56SSGKK265 pKa = 10.06VVGGLDD271 pKa = 3.6NRR273 pKa = 11.84VSPYY277 pKa = 9.64TGNSGDD283 pKa = 4.18TPTMQCDD290 pKa = 3.55MLQLKK295 pKa = 9.31PGRR298 pKa = 11.84YY299 pKa = 8.55SVRR302 pKa = 11.84SSPRR306 pKa = 11.84FLLMPEE312 pKa = 4.1RR313 pKa = 11.84SYY315 pKa = 11.73CFDD318 pKa = 3.5MKK320 pKa = 10.99EE321 pKa = 4.16KK322 pKa = 11.04GPVTAVQSIWGKK334 pKa = 10.35GRR336 pKa = 11.84EE337 pKa = 4.07SDD339 pKa = 3.62HH340 pKa = 7.93AVDD343 pKa = 4.18QACLSTPGCMLIQKK357 pKa = 7.37QKK359 pKa = 10.35PYY361 pKa = 10.24IGEE364 pKa = 4.33ADD366 pKa = 3.72DD367 pKa = 3.83HH368 pKa = 7.68HH369 pKa = 7.57GDD371 pKa = 3.26QEE373 pKa = 4.19MRR375 pKa = 11.84EE376 pKa = 4.26LLSGLDD382 pKa = 3.52YY383 pKa = 10.79EE384 pKa = 5.02ARR386 pKa = 11.84CISQSGWVNEE396 pKa = 4.0TSPFTEE402 pKa = 5.12EE403 pKa = 3.88YY404 pKa = 10.82LLPPKK409 pKa = 10.63FGRR412 pKa = 11.84CPLAAKK418 pKa = 10.04EE419 pKa = 3.87EE420 pKa = 4.69SIPKK424 pKa = 9.95IPDD427 pKa = 3.22GLLIPTSGTDD437 pKa = 3.31TTVTKK442 pKa = 10.09PKK444 pKa = 10.63SRR446 pKa = 11.84IFGIDD451 pKa = 3.38DD452 pKa = 5.09LIIGLLFVAIVEE464 pKa = 4.27AGIGGYY470 pKa = 10.53LLGSRR475 pKa = 11.84KK476 pKa = 9.78VSGGGVTKK484 pKa = 10.59EE485 pKa = 3.9SAEE488 pKa = 4.1KK489 pKa = 10.63GFEE492 pKa = 4.3KK493 pKa = 10.09IGNDD497 pKa = 3.11IQILRR502 pKa = 11.84SSTNIAIEE510 pKa = 4.02KK511 pKa = 9.93LNDD514 pKa = 4.38RR515 pKa = 11.84ISHH518 pKa = 6.87DD519 pKa = 3.58EE520 pKa = 3.67QAIRR524 pKa = 11.84DD525 pKa = 3.81LTLEE529 pKa = 4.02IEE531 pKa = 4.48NARR534 pKa = 11.84SEE536 pKa = 4.27ALLGEE541 pKa = 4.29LGIIRR546 pKa = 11.84ALLVGNISIGLQEE559 pKa = 4.78SLWEE563 pKa = 3.97LASEE567 pKa = 4.14ITNRR571 pKa = 11.84AGDD574 pKa = 3.74LAVEE578 pKa = 4.25VSPGCWVIDD587 pKa = 4.49NNICDD592 pKa = 3.64QSCQNFIFKK601 pKa = 10.49FNEE604 pKa = 3.92TAPVPTIPPLDD615 pKa = 3.72TKK617 pKa = 10.71IDD619 pKa = 3.76LQSDD623 pKa = 4.09PFYY626 pKa = 10.2WGSSLGLAITAAISLAALVISGIAICRR653 pKa = 11.84TKK655 pKa = 11.06
Molecular weight: 71.96 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q6I7C2|PA_INCAA Polymerase acidic protein OS=Influenza C virus (strain C/Ann Arbor/1/1950) OX=11553 GN=PA PE=3 SV=1
MM1 pKa = 7.83SDD3 pKa = 2.89RR4 pKa = 11.84RR5 pKa = 11.84QNRR8 pKa = 11.84KK9 pKa = 7.56TPDD12 pKa = 3.24EE13 pKa = 3.94QRR15 pKa = 11.84KK16 pKa = 9.78ANALIINEE24 pKa = 4.26NIEE27 pKa = 4.08AYY29 pKa = 9.61IAICKK34 pKa = 9.53EE35 pKa = 3.66VGLNGDD41 pKa = 3.56EE42 pKa = 4.26MLILEE47 pKa = 4.5NGIAIEE53 pKa = 3.93KK54 pKa = 10.08AIRR57 pKa = 11.84ICCDD61 pKa = 2.38GKK63 pKa = 10.13YY64 pKa = 9.29QEE66 pKa = 4.64KK67 pKa = 10.08RR68 pKa = 11.84EE69 pKa = 4.12KK70 pKa = 10.04KK71 pKa = 9.88ARR73 pKa = 11.84EE74 pKa = 3.88AQRR77 pKa = 11.84ADD79 pKa = 3.47SNFNADD85 pKa = 3.52SIGIRR90 pKa = 11.84LVKK93 pKa = 10.35RR94 pKa = 11.84AGSGTNITYY103 pKa = 9.98HH104 pKa = 6.27AVVEE108 pKa = 4.27LTSRR112 pKa = 11.84SRR114 pKa = 11.84IVQILKK120 pKa = 8.9SHH122 pKa = 7.09WGNEE126 pKa = 3.72LNRR129 pKa = 11.84AKK131 pKa = 10.48IAGKK135 pKa = 10.23RR136 pKa = 11.84LGFSALFASNLEE148 pKa = 4.16AIIYY152 pKa = 9.11QRR154 pKa = 11.84GRR156 pKa = 11.84NAARR160 pKa = 11.84RR161 pKa = 11.84NGSAEE166 pKa = 4.46LFTLTQGAGIEE177 pKa = 4.19TRR179 pKa = 11.84YY180 pKa = 9.47KK181 pKa = 10.06WIMEE185 pKa = 3.75KK186 pKa = 10.51HH187 pKa = 5.69IGIGVLIADD196 pKa = 4.58AKK198 pKa = 11.06GLINGKK204 pKa = 9.56RR205 pKa = 11.84EE206 pKa = 3.92GKK208 pKa = 10.23RR209 pKa = 11.84GVDD212 pKa = 3.21ANVKK216 pKa = 9.73LRR218 pKa = 11.84AGTTGSPLEE227 pKa = 4.09RR228 pKa = 11.84AMQGIEE234 pKa = 3.93KK235 pKa = 10.25KK236 pKa = 10.62AFPGPLRR243 pKa = 11.84ALARR247 pKa = 11.84RR248 pKa = 11.84VVKK251 pKa = 10.75ANYY254 pKa = 9.66NDD256 pKa = 3.09ARR258 pKa = 11.84EE259 pKa = 4.19ALNVIAEE266 pKa = 4.22ASLLLKK272 pKa = 10.13PQITNKK278 pKa = 7.67MTMPWCMWLAARR290 pKa = 11.84LTLKK294 pKa = 11.01DD295 pKa = 3.52EE296 pKa = 4.41FANFCAYY303 pKa = 10.19AGRR306 pKa = 11.84RR307 pKa = 11.84AFEE310 pKa = 4.07VFNIAMEE317 pKa = 4.64KK318 pKa = 10.09IGICSFQGTIMNDD331 pKa = 3.39DD332 pKa = 4.45EE333 pKa = 5.4IEE335 pKa = 4.41SIEE338 pKa = 4.93DD339 pKa = 3.31KK340 pKa = 11.27AQVLMMACFGLAYY353 pKa = 9.94EE354 pKa = 4.84DD355 pKa = 4.71FSLVSAMVSHH365 pKa = 7.32PLKK368 pKa = 10.38LRR370 pKa = 11.84NRR372 pKa = 11.84MKK374 pKa = 10.24IGNFRR379 pKa = 11.84VGEE382 pKa = 4.17KK383 pKa = 10.52VSTVLSPLLRR393 pKa = 11.84FTRR396 pKa = 11.84WAEE399 pKa = 3.67FAQRR403 pKa = 11.84FALQANTSRR412 pKa = 11.84EE413 pKa = 3.87GAQISNSAVFAVEE426 pKa = 4.41RR427 pKa = 11.84KK428 pKa = 8.24ITTDD432 pKa = 3.15VQRR435 pKa = 11.84VEE437 pKa = 4.06EE438 pKa = 4.46LLNKK442 pKa = 8.94VQAHH446 pKa = 5.9EE447 pKa = 4.75DD448 pKa = 3.71EE449 pKa = 4.97PLQTLYY455 pKa = 11.16KK456 pKa = 9.96KK457 pKa = 10.59VRR459 pKa = 11.84EE460 pKa = 4.02QISIIGRR467 pKa = 11.84NKK469 pKa = 10.34SEE471 pKa = 3.73IKK473 pKa = 10.41EE474 pKa = 4.03FLGSSMYY481 pKa = 10.88DD482 pKa = 3.13LNDD485 pKa = 3.36QEE487 pKa = 4.62KK488 pKa = 10.41QNPINFRR495 pKa = 11.84SGAHH499 pKa = 5.88PFFFEE504 pKa = 5.07FDD506 pKa = 3.53PDD508 pKa = 3.82YY509 pKa = 11.6NPIRR513 pKa = 11.84VKK515 pKa = 10.61RR516 pKa = 11.84PKK518 pKa = 10.57KK519 pKa = 10.15PIAKK523 pKa = 9.26RR524 pKa = 11.84NSNISRR530 pKa = 11.84LEE532 pKa = 3.86EE533 pKa = 3.73EE534 pKa = 5.15GMDD537 pKa = 3.66EE538 pKa = 4.51NSEE541 pKa = 3.74IGQAKK546 pKa = 9.44KK547 pKa = 9.97MKK549 pKa = 9.96PLDD552 pKa = 3.68QLTSTSSNIPGKK564 pKa = 10.28NN565 pKa = 3.02
MM1 pKa = 7.83SDD3 pKa = 2.89RR4 pKa = 11.84RR5 pKa = 11.84QNRR8 pKa = 11.84KK9 pKa = 7.56TPDD12 pKa = 3.24EE13 pKa = 3.94QRR15 pKa = 11.84KK16 pKa = 9.78ANALIINEE24 pKa = 4.26NIEE27 pKa = 4.08AYY29 pKa = 9.61IAICKK34 pKa = 9.53EE35 pKa = 3.66VGLNGDD41 pKa = 3.56EE42 pKa = 4.26MLILEE47 pKa = 4.5NGIAIEE53 pKa = 3.93KK54 pKa = 10.08AIRR57 pKa = 11.84ICCDD61 pKa = 2.38GKK63 pKa = 10.13YY64 pKa = 9.29QEE66 pKa = 4.64KK67 pKa = 10.08RR68 pKa = 11.84EE69 pKa = 4.12KK70 pKa = 10.04KK71 pKa = 9.88ARR73 pKa = 11.84EE74 pKa = 3.88AQRR77 pKa = 11.84ADD79 pKa = 3.47SNFNADD85 pKa = 3.52SIGIRR90 pKa = 11.84LVKK93 pKa = 10.35RR94 pKa = 11.84AGSGTNITYY103 pKa = 9.98HH104 pKa = 6.27AVVEE108 pKa = 4.27LTSRR112 pKa = 11.84SRR114 pKa = 11.84IVQILKK120 pKa = 8.9SHH122 pKa = 7.09WGNEE126 pKa = 3.72LNRR129 pKa = 11.84AKK131 pKa = 10.48IAGKK135 pKa = 10.23RR136 pKa = 11.84LGFSALFASNLEE148 pKa = 4.16AIIYY152 pKa = 9.11QRR154 pKa = 11.84GRR156 pKa = 11.84NAARR160 pKa = 11.84RR161 pKa = 11.84NGSAEE166 pKa = 4.46LFTLTQGAGIEE177 pKa = 4.19TRR179 pKa = 11.84YY180 pKa = 9.47KK181 pKa = 10.06WIMEE185 pKa = 3.75KK186 pKa = 10.51HH187 pKa = 5.69IGIGVLIADD196 pKa = 4.58AKK198 pKa = 11.06GLINGKK204 pKa = 9.56RR205 pKa = 11.84EE206 pKa = 3.92GKK208 pKa = 10.23RR209 pKa = 11.84GVDD212 pKa = 3.21ANVKK216 pKa = 9.73LRR218 pKa = 11.84AGTTGSPLEE227 pKa = 4.09RR228 pKa = 11.84AMQGIEE234 pKa = 3.93KK235 pKa = 10.25KK236 pKa = 10.62AFPGPLRR243 pKa = 11.84ALARR247 pKa = 11.84RR248 pKa = 11.84VVKK251 pKa = 10.75ANYY254 pKa = 9.66NDD256 pKa = 3.09ARR258 pKa = 11.84EE259 pKa = 4.19ALNVIAEE266 pKa = 4.22ASLLLKK272 pKa = 10.13PQITNKK278 pKa = 7.67MTMPWCMWLAARR290 pKa = 11.84LTLKK294 pKa = 11.01DD295 pKa = 3.52EE296 pKa = 4.41FANFCAYY303 pKa = 10.19AGRR306 pKa = 11.84RR307 pKa = 11.84AFEE310 pKa = 4.07VFNIAMEE317 pKa = 4.64KK318 pKa = 10.09IGICSFQGTIMNDD331 pKa = 3.39DD332 pKa = 4.45EE333 pKa = 5.4IEE335 pKa = 4.41SIEE338 pKa = 4.93DD339 pKa = 3.31KK340 pKa = 11.27AQVLMMACFGLAYY353 pKa = 9.94EE354 pKa = 4.84DD355 pKa = 4.71FSLVSAMVSHH365 pKa = 7.32PLKK368 pKa = 10.38LRR370 pKa = 11.84NRR372 pKa = 11.84MKK374 pKa = 10.24IGNFRR379 pKa = 11.84VGEE382 pKa = 4.17KK383 pKa = 10.52VSTVLSPLLRR393 pKa = 11.84FTRR396 pKa = 11.84WAEE399 pKa = 3.67FAQRR403 pKa = 11.84FALQANTSRR412 pKa = 11.84EE413 pKa = 3.87GAQISNSAVFAVEE426 pKa = 4.41RR427 pKa = 11.84KK428 pKa = 8.24ITTDD432 pKa = 3.15VQRR435 pKa = 11.84VEE437 pKa = 4.06EE438 pKa = 4.46LLNKK442 pKa = 8.94VQAHH446 pKa = 5.9EE447 pKa = 4.75DD448 pKa = 3.71EE449 pKa = 4.97PLQTLYY455 pKa = 11.16KK456 pKa = 9.96KK457 pKa = 10.59VRR459 pKa = 11.84EE460 pKa = 4.02QISIIGRR467 pKa = 11.84NKK469 pKa = 10.34SEE471 pKa = 3.73IKK473 pKa = 10.41EE474 pKa = 4.03FLGSSMYY481 pKa = 10.88DD482 pKa = 3.13LNDD485 pKa = 3.36QEE487 pKa = 4.62KK488 pKa = 10.41QNPINFRR495 pKa = 11.84SGAHH499 pKa = 5.88PFFFEE504 pKa = 5.07FDD506 pKa = 3.53PDD508 pKa = 3.82YY509 pKa = 11.6NPIRR513 pKa = 11.84VKK515 pKa = 10.61RR516 pKa = 11.84PKK518 pKa = 10.57KK519 pKa = 10.15PIAKK523 pKa = 9.26RR524 pKa = 11.84NSNISRR530 pKa = 11.84LEE532 pKa = 3.86EE533 pKa = 3.73EE534 pKa = 5.15GMDD537 pKa = 3.66EE538 pKa = 4.51NSEE541 pKa = 3.74IGQAKK546 pKa = 9.44KK547 pKa = 9.97MKK549 pKa = 9.96PLDD552 pKa = 3.68QLTSTSSNIPGKK564 pKa = 10.28NN565 pKa = 3.02
Molecular weight: 63.63 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
4501 |
182 |
774 |
500.1 |
56.51 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.91 ± 0.553 | 2.155 ± 0.236 |
4.754 ± 0.191 | 7.976 ± 0.496 |
4.177 ± 0.409 | 6.043 ± 0.472 |
1.4 ± 0.149 | 7.087 ± 0.323 |
8.443 ± 0.438 | 8.643 ± 0.425 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.955 ± 0.508 | 4.91 ± 0.326 |
3.866 ± 0.252 | 2.977 ± 0.304 |
5.643 ± 0.506 | 6.976 ± 0.512 |
5.554 ± 0.436 | 4.888 ± 0.336 |
1.178 ± 0.08 | 2.466 ± 0.204 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
