
Auritidibacter sp. NML120636
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Micrococcaceae; Auritidibacter; unclassified Auritidibacter
Average proteome isoelectric point is 5.83
Get precalculated fractions of proteins
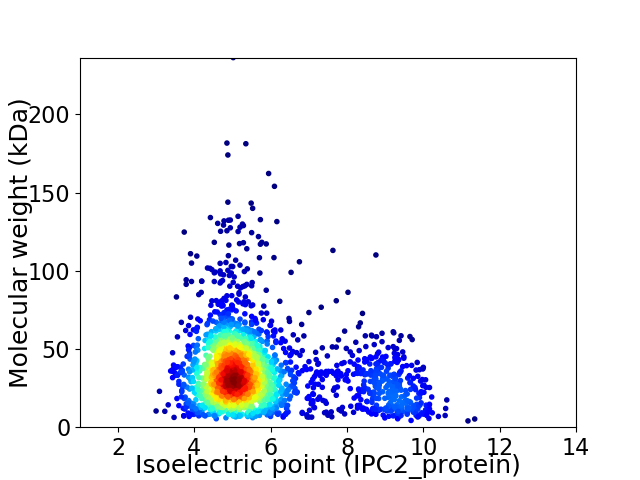
Virtual 2D-PAGE plot for 2118 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A318AH37|A0A318AH37_9MICC C4-dicarboxylate ABC transporter substrate-binding protein OS=Auritidibacter sp. NML120636 OX=2170743 GN=DCC25_09155 PE=4 SV=1
MM1 pKa = 7.66ALTGSHH7 pKa = 6.77HH8 pKa = 5.89EE9 pKa = 4.24HH10 pKa = 6.83SLTTSLTRR18 pKa = 11.84YY19 pKa = 9.03FVGFATLLLLLTGCAGSGDD38 pKa = 4.15PEE40 pKa = 4.39AADD43 pKa = 3.59TSSIPGTDD51 pKa = 3.22TQDD54 pKa = 3.25APEE57 pKa = 4.99PPTEE61 pKa = 4.97AEE63 pKa = 4.68DD64 pKa = 3.81SSSAPDD70 pKa = 3.59TPSTEE75 pKa = 4.36EE76 pKa = 3.99SSEE79 pKa = 4.09EE80 pKa = 3.9PPEE83 pKa = 4.04PTEE86 pKa = 4.07PEE88 pKa = 3.99IEE90 pKa = 4.29SLEE93 pKa = 4.21DD94 pKa = 3.28TDD96 pKa = 4.83FSSISFQFVGYY107 pKa = 10.48SDD109 pKa = 3.93TDD111 pKa = 3.52DD112 pKa = 4.93SEE114 pKa = 4.76EE115 pKa = 3.97KK116 pKa = 10.22TFVDD120 pKa = 4.21GTVEE124 pKa = 4.04AGDD127 pKa = 3.53PQFPVTYY134 pKa = 9.56EE135 pKa = 3.79LGEE138 pKa = 4.6VIHH141 pKa = 6.71QDD143 pKa = 3.38LNGDD147 pKa = 4.37GIVDD151 pKa = 3.58AAVEE155 pKa = 3.95IEE157 pKa = 4.16RR158 pKa = 11.84YY159 pKa = 9.32EE160 pKa = 4.23GNSYY164 pKa = 10.82DD165 pKa = 3.64GRR167 pKa = 11.84WYY169 pKa = 9.53MWIATEE175 pKa = 5.14DD176 pKa = 3.94GPEE179 pKa = 3.92QSPLPFARR187 pKa = 11.84SGNCGDD193 pKa = 3.54QVEE196 pKa = 5.03SVTATNDD203 pKa = 3.12GLEE206 pKa = 4.21VHH208 pKa = 5.96EE209 pKa = 4.62HH210 pKa = 5.66LRR212 pKa = 11.84AKK214 pKa = 10.44EE215 pKa = 3.98DD216 pKa = 3.67DD217 pKa = 4.05VSCAEE222 pKa = 4.4HH223 pKa = 6.56GLHH226 pKa = 6.31EE227 pKa = 4.36RR228 pKa = 11.84TRR230 pKa = 11.84TIAADD235 pKa = 3.69YY236 pKa = 10.61FDD238 pKa = 5.88LAGAWWPVQVAPVIGYY254 pKa = 10.11GGICSLTGDD263 pKa = 4.19EE264 pKa = 5.6GMTAPVDD271 pKa = 3.48IAHH274 pKa = 5.88VNPYY278 pKa = 9.93KK279 pKa = 10.57EE280 pKa = 4.26SASIAEE286 pKa = 4.66TNAEE290 pKa = 4.19THH292 pKa = 6.83PEE294 pKa = 3.95LTILFPFGEE303 pKa = 4.79GDD305 pKa = 3.43GSDD308 pKa = 3.22RR309 pKa = 11.84KK310 pKa = 10.01IDD312 pKa = 3.84GEE314 pKa = 3.81WWAIGGIFDD323 pKa = 4.22GSQVLGCGWVAYY335 pKa = 9.38EE336 pKa = 4.92DD337 pKa = 4.09LPIGIRR343 pKa = 11.84FGG345 pKa = 3.44
MM1 pKa = 7.66ALTGSHH7 pKa = 6.77HH8 pKa = 5.89EE9 pKa = 4.24HH10 pKa = 6.83SLTTSLTRR18 pKa = 11.84YY19 pKa = 9.03FVGFATLLLLLTGCAGSGDD38 pKa = 4.15PEE40 pKa = 4.39AADD43 pKa = 3.59TSSIPGTDD51 pKa = 3.22TQDD54 pKa = 3.25APEE57 pKa = 4.99PPTEE61 pKa = 4.97AEE63 pKa = 4.68DD64 pKa = 3.81SSSAPDD70 pKa = 3.59TPSTEE75 pKa = 4.36EE76 pKa = 3.99SSEE79 pKa = 4.09EE80 pKa = 3.9PPEE83 pKa = 4.04PTEE86 pKa = 4.07PEE88 pKa = 3.99IEE90 pKa = 4.29SLEE93 pKa = 4.21DD94 pKa = 3.28TDD96 pKa = 4.83FSSISFQFVGYY107 pKa = 10.48SDD109 pKa = 3.93TDD111 pKa = 3.52DD112 pKa = 4.93SEE114 pKa = 4.76EE115 pKa = 3.97KK116 pKa = 10.22TFVDD120 pKa = 4.21GTVEE124 pKa = 4.04AGDD127 pKa = 3.53PQFPVTYY134 pKa = 9.56EE135 pKa = 3.79LGEE138 pKa = 4.6VIHH141 pKa = 6.71QDD143 pKa = 3.38LNGDD147 pKa = 4.37GIVDD151 pKa = 3.58AAVEE155 pKa = 3.95IEE157 pKa = 4.16RR158 pKa = 11.84YY159 pKa = 9.32EE160 pKa = 4.23GNSYY164 pKa = 10.82DD165 pKa = 3.64GRR167 pKa = 11.84WYY169 pKa = 9.53MWIATEE175 pKa = 5.14DD176 pKa = 3.94GPEE179 pKa = 3.92QSPLPFARR187 pKa = 11.84SGNCGDD193 pKa = 3.54QVEE196 pKa = 5.03SVTATNDD203 pKa = 3.12GLEE206 pKa = 4.21VHH208 pKa = 5.96EE209 pKa = 4.62HH210 pKa = 5.66LRR212 pKa = 11.84AKK214 pKa = 10.44EE215 pKa = 3.98DD216 pKa = 3.67DD217 pKa = 4.05VSCAEE222 pKa = 4.4HH223 pKa = 6.56GLHH226 pKa = 6.31EE227 pKa = 4.36RR228 pKa = 11.84TRR230 pKa = 11.84TIAADD235 pKa = 3.69YY236 pKa = 10.61FDD238 pKa = 5.88LAGAWWPVQVAPVIGYY254 pKa = 10.11GGICSLTGDD263 pKa = 4.19EE264 pKa = 5.6GMTAPVDD271 pKa = 3.48IAHH274 pKa = 5.88VNPYY278 pKa = 9.93KK279 pKa = 10.57EE280 pKa = 4.26SASIAEE286 pKa = 4.66TNAEE290 pKa = 4.19THH292 pKa = 6.83PEE294 pKa = 3.95LTILFPFGEE303 pKa = 4.79GDD305 pKa = 3.43GSDD308 pKa = 3.22RR309 pKa = 11.84KK310 pKa = 10.01IDD312 pKa = 3.84GEE314 pKa = 3.81WWAIGGIFDD323 pKa = 4.22GSQVLGCGWVAYY335 pKa = 9.38EE336 pKa = 4.92DD337 pKa = 4.09LPIGIRR343 pKa = 11.84FGG345 pKa = 3.44
Molecular weight: 37.09 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A318AGA3|A0A318AGA3_9MICC Uncharacterized protein OS=Auritidibacter sp. NML120636 OX=2170743 GN=DCC25_00390 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84ARR15 pKa = 11.84KK16 pKa = 8.24HH17 pKa = 4.59GFRR20 pKa = 11.84ARR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AVLSARR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.31GRR40 pKa = 11.84ARR42 pKa = 11.84VSAA45 pKa = 4.0
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84ARR15 pKa = 11.84KK16 pKa = 8.24HH17 pKa = 4.59GFRR20 pKa = 11.84ARR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AVLSARR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.31GRR40 pKa = 11.84ARR42 pKa = 11.84VSAA45 pKa = 4.0
Molecular weight: 5.25 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
715878 |
31 |
2147 |
338.0 |
36.76 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.97 ± 0.052 | 0.593 ± 0.013 |
6.149 ± 0.045 | 6.377 ± 0.049 |
3.087 ± 0.031 | 8.009 ± 0.045 |
2.558 ± 0.029 | 4.907 ± 0.035 |
2.515 ± 0.042 | 9.797 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.115 ± 0.024 | 2.577 ± 0.023 |
5.214 ± 0.035 | 4.21 ± 0.035 |
6.348 ± 0.051 | 6.11 ± 0.042 |
6.627 ± 0.037 | 8.199 ± 0.046 |
1.408 ± 0.022 | 2.23 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
