
Roseimaritima ulvae
Taxonomy: cellular organisms; Bacteria; PVC group; Planctomycetes; Planctomycetia; Pirellulales; Pirellulaceae; Roseimaritima
Average proteome isoelectric point is 6.12
Get precalculated fractions of proteins
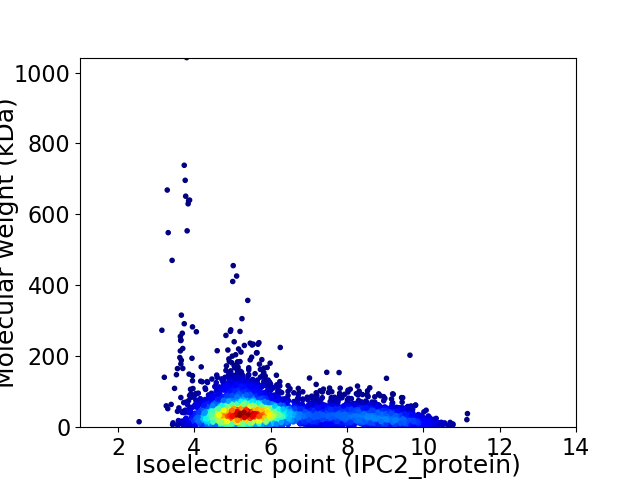
Virtual 2D-PAGE plot for 5779 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5B9QM57|A0A5B9QM57_9BACT Sensory transduction protein regX3 OS=Roseimaritima ulvae OX=980254 GN=regX3_2 PE=4 SV=1
MM1 pKa = 7.22PTLPRR6 pKa = 11.84HH7 pKa = 5.52RR8 pKa = 11.84RR9 pKa = 11.84SLRR12 pKa = 11.84CEE14 pKa = 3.63SLEE17 pKa = 4.43RR18 pKa = 11.84RR19 pKa = 11.84NLLAAGFGLEE29 pKa = 4.54HH30 pKa = 6.71NFISPEE36 pKa = 4.01DD37 pKa = 3.76VDD39 pKa = 4.32GSGDD43 pKa = 3.57VTPYY47 pKa = 10.96DD48 pKa = 3.51ALQVVNNLNRR58 pKa = 11.84QARR61 pKa = 11.84SEE63 pKa = 4.14GQGEE67 pKa = 4.38DD68 pKa = 3.47SSSSSGLMLDD78 pKa = 3.62VDD80 pKa = 4.48ADD82 pKa = 4.24GTSSPSDD89 pKa = 2.57AWRR92 pKa = 11.84IINYY96 pKa = 9.39LNQNGVHH103 pKa = 6.27GALASSLVPIHH114 pKa = 7.02DD115 pKa = 5.01RR116 pKa = 11.84IAALEE121 pKa = 4.19SALHH125 pKa = 5.96SSSVPSWLGSGTAHH139 pKa = 7.95DD140 pKa = 4.73ILASLYY146 pKa = 10.79NGIAPEE152 pKa = 4.27INSDD156 pKa = 4.04LQDD159 pKa = 3.92EE160 pKa = 4.88LDD162 pKa = 4.05DD163 pKa = 4.22ALEE166 pKa = 4.34RR167 pKa = 11.84FDD169 pKa = 6.22LQLQYY174 pKa = 11.57NEE176 pKa = 4.4LSDD179 pKa = 4.16RR180 pKa = 11.84LDD182 pKa = 4.2SIADD186 pKa = 3.89DD187 pKa = 4.48LPSFDD192 pKa = 6.0SIFSQLGSQIGSQYY206 pKa = 10.99HH207 pKa = 5.42EE208 pKa = 4.84LQDD211 pKa = 3.78EE212 pKa = 4.32LADD215 pKa = 4.05ALEE218 pKa = 4.36EE219 pKa = 4.06LHH221 pKa = 7.07DD222 pKa = 3.95QAGSVLDD229 pKa = 3.98EE230 pKa = 5.08LFDD233 pKa = 3.64DD234 pKa = 5.45HH235 pKa = 8.9YY236 pKa = 11.8YY237 pKa = 11.29DD238 pKa = 6.1DD239 pKa = 6.18DD240 pKa = 5.03SDD242 pKa = 4.32EE243 pKa = 4.24NTEE246 pKa = 4.34SVNGLAEE253 pKa = 4.46NGLSDD258 pKa = 5.83DD259 pKa = 5.07DD260 pKa = 5.37DD261 pKa = 5.44SSTTEE266 pKa = 3.68QVSDD270 pKa = 3.73VLSHH274 pKa = 7.44IDD276 pKa = 3.7EE277 pKa = 4.31EE278 pKa = 4.91LEE280 pKa = 3.84NLFYY284 pKa = 11.16DD285 pKa = 4.16VASDD289 pKa = 3.13IHH291 pKa = 6.9DD292 pKa = 4.29LNFSGVLNTLANVVHH307 pKa = 5.66VHH309 pKa = 6.61LPLLSSGLPDD319 pKa = 4.74LFDD322 pKa = 5.08SNIVDD327 pKa = 3.54IPNDD331 pKa = 3.61FSSEE335 pKa = 3.91LSDD338 pKa = 4.39LYY340 pKa = 11.45DD341 pKa = 3.83RR342 pKa = 11.84LEE344 pKa = 3.98SVYY347 pKa = 11.2DD348 pKa = 3.65DD349 pKa = 3.58VVPEE353 pKa = 4.19VEE355 pKa = 4.59DD356 pKa = 4.21ALDD359 pKa = 3.46HH360 pKa = 6.41VFDD363 pKa = 3.87SFSYY367 pKa = 9.25IGSAASHH374 pKa = 6.64LSSFVHH380 pKa = 5.81YY381 pKa = 9.97LHH383 pKa = 7.61GYY385 pKa = 8.54GSDD388 pKa = 3.65YY389 pKa = 10.78NTATT393 pKa = 3.68
MM1 pKa = 7.22PTLPRR6 pKa = 11.84HH7 pKa = 5.52RR8 pKa = 11.84RR9 pKa = 11.84SLRR12 pKa = 11.84CEE14 pKa = 3.63SLEE17 pKa = 4.43RR18 pKa = 11.84RR19 pKa = 11.84NLLAAGFGLEE29 pKa = 4.54HH30 pKa = 6.71NFISPEE36 pKa = 4.01DD37 pKa = 3.76VDD39 pKa = 4.32GSGDD43 pKa = 3.57VTPYY47 pKa = 10.96DD48 pKa = 3.51ALQVVNNLNRR58 pKa = 11.84QARR61 pKa = 11.84SEE63 pKa = 4.14GQGEE67 pKa = 4.38DD68 pKa = 3.47SSSSSGLMLDD78 pKa = 3.62VDD80 pKa = 4.48ADD82 pKa = 4.24GTSSPSDD89 pKa = 2.57AWRR92 pKa = 11.84IINYY96 pKa = 9.39LNQNGVHH103 pKa = 6.27GALASSLVPIHH114 pKa = 7.02DD115 pKa = 5.01RR116 pKa = 11.84IAALEE121 pKa = 4.19SALHH125 pKa = 5.96SSSVPSWLGSGTAHH139 pKa = 7.95DD140 pKa = 4.73ILASLYY146 pKa = 10.79NGIAPEE152 pKa = 4.27INSDD156 pKa = 4.04LQDD159 pKa = 3.92EE160 pKa = 4.88LDD162 pKa = 4.05DD163 pKa = 4.22ALEE166 pKa = 4.34RR167 pKa = 11.84FDD169 pKa = 6.22LQLQYY174 pKa = 11.57NEE176 pKa = 4.4LSDD179 pKa = 4.16RR180 pKa = 11.84LDD182 pKa = 4.2SIADD186 pKa = 3.89DD187 pKa = 4.48LPSFDD192 pKa = 6.0SIFSQLGSQIGSQYY206 pKa = 10.99HH207 pKa = 5.42EE208 pKa = 4.84LQDD211 pKa = 3.78EE212 pKa = 4.32LADD215 pKa = 4.05ALEE218 pKa = 4.36EE219 pKa = 4.06LHH221 pKa = 7.07DD222 pKa = 3.95QAGSVLDD229 pKa = 3.98EE230 pKa = 5.08LFDD233 pKa = 3.64DD234 pKa = 5.45HH235 pKa = 8.9YY236 pKa = 11.8YY237 pKa = 11.29DD238 pKa = 6.1DD239 pKa = 6.18DD240 pKa = 5.03SDD242 pKa = 4.32EE243 pKa = 4.24NTEE246 pKa = 4.34SVNGLAEE253 pKa = 4.46NGLSDD258 pKa = 5.83DD259 pKa = 5.07DD260 pKa = 5.37DD261 pKa = 5.44SSTTEE266 pKa = 3.68QVSDD270 pKa = 3.73VLSHH274 pKa = 7.44IDD276 pKa = 3.7EE277 pKa = 4.31EE278 pKa = 4.91LEE280 pKa = 3.84NLFYY284 pKa = 11.16DD285 pKa = 4.16VASDD289 pKa = 3.13IHH291 pKa = 6.9DD292 pKa = 4.29LNFSGVLNTLANVVHH307 pKa = 5.66VHH309 pKa = 6.61LPLLSSGLPDD319 pKa = 4.74LFDD322 pKa = 5.08SNIVDD327 pKa = 3.54IPNDD331 pKa = 3.61FSSEE335 pKa = 3.91LSDD338 pKa = 4.39LYY340 pKa = 11.45DD341 pKa = 3.83RR342 pKa = 11.84LEE344 pKa = 3.98SVYY347 pKa = 11.2DD348 pKa = 3.65DD349 pKa = 3.58VVPEE353 pKa = 4.19VEE355 pKa = 4.59DD356 pKa = 4.21ALDD359 pKa = 3.46HH360 pKa = 6.41VFDD363 pKa = 3.87SFSYY367 pKa = 9.25IGSAASHH374 pKa = 6.64LSSFVHH380 pKa = 5.81YY381 pKa = 9.97LHH383 pKa = 7.61GYY385 pKa = 8.54GSDD388 pKa = 3.65YY389 pKa = 10.78NTATT393 pKa = 3.68
Molecular weight: 43.06 kDa
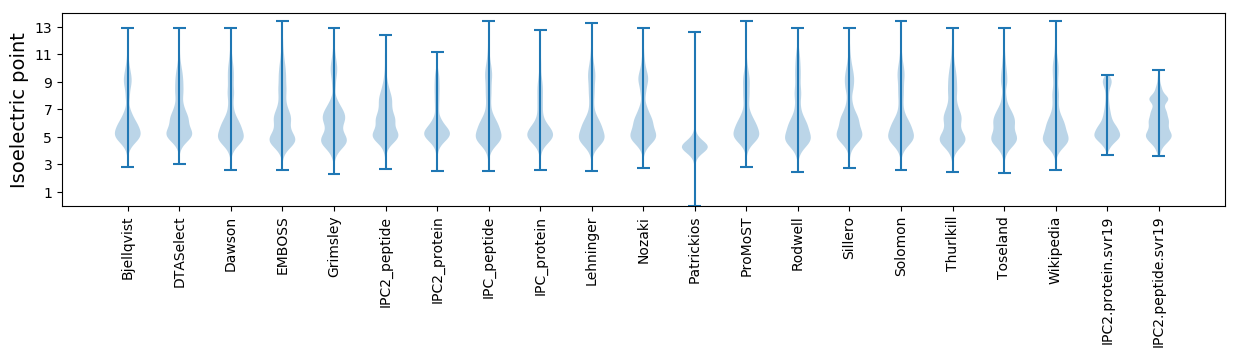
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5B9QKU1|A0A5B9QKU1_9BACT 50S ribosomal protein L7/L12 OS=Roseimaritima ulvae OX=980254 GN=rplL_1 PE=3 SV=1
MM1 pKa = 7.1VRR3 pKa = 11.84LSRR6 pKa = 11.84LTRR9 pKa = 11.84SRR11 pKa = 11.84RR12 pKa = 11.84SLKK15 pKa = 10.24KK16 pKa = 10.48FRR18 pKa = 11.84THH20 pKa = 4.99PHH22 pKa = 5.91ARR24 pKa = 11.84QAPQAQAWSAGRR36 pKa = 11.84SQSKK40 pKa = 10.44PNGLEE45 pKa = 3.92KK46 pKa = 10.8QKK48 pKa = 9.96TNPTLRR54 pKa = 11.84GSPSRR59 pKa = 11.84RR60 pKa = 11.84LRR62 pKa = 11.84LRR64 pKa = 11.84VKK66 pKa = 10.34RR67 pKa = 11.84GSLRR71 pKa = 11.84SLKK74 pKa = 10.32KK75 pKa = 10.54FRR77 pKa = 11.84THH79 pKa = 4.99PHH81 pKa = 5.91ARR83 pKa = 11.84QAPQAQAWSAGRR95 pKa = 11.84SQSKK99 pKa = 10.44PNGLEE104 pKa = 3.92KK105 pKa = 10.8QKK107 pKa = 9.96TNPTLRR113 pKa = 11.84GSPRR117 pKa = 11.84RR118 pKa = 11.84RR119 pKa = 11.84LRR121 pKa = 11.84LRR123 pKa = 11.84VKK125 pKa = 9.95RR126 pKa = 11.84GSRR129 pKa = 11.84RR130 pKa = 11.84SLKK133 pKa = 10.27KK134 pKa = 10.52FRR136 pKa = 11.84THH138 pKa = 4.99PHH140 pKa = 5.91ARR142 pKa = 11.84QAPQAQAWSAGRR154 pKa = 11.84SQSKK158 pKa = 10.44PNGLEE163 pKa = 3.92KK164 pKa = 10.8QKK166 pKa = 9.96TNPTLRR172 pKa = 11.84GSPRR176 pKa = 11.84RR177 pKa = 11.84RR178 pKa = 11.84LRR180 pKa = 11.84LRR182 pKa = 11.84VKK184 pKa = 9.95RR185 pKa = 11.84GSRR188 pKa = 11.84RR189 pKa = 11.84SLKK192 pKa = 10.27KK193 pKa = 10.52FRR195 pKa = 11.84THH197 pKa = 4.99PHH199 pKa = 5.91ARR201 pKa = 11.84QAPQAQAWSAGRR213 pKa = 11.84SQSKK217 pKa = 10.44PNGLEE222 pKa = 3.92KK223 pKa = 10.8QKK225 pKa = 9.96TNPTLRR231 pKa = 11.84GSPRR235 pKa = 11.84RR236 pKa = 11.84RR237 pKa = 11.84LRR239 pKa = 11.84LRR241 pKa = 11.84VKK243 pKa = 9.93RR244 pKa = 11.84DD245 pKa = 2.99SRR247 pKa = 11.84RR248 pKa = 11.84SLKK251 pKa = 10.4KK252 pKa = 10.21FHH254 pKa = 5.99THH256 pKa = 4.19PHH258 pKa = 5.8ARR260 pKa = 11.84QAPQAQAWSAGRR272 pKa = 11.84SQSKK276 pKa = 10.44PNGLEE281 pKa = 3.92KK282 pKa = 10.8QKK284 pKa = 9.96TNPTLRR290 pKa = 11.84GSPRR294 pKa = 11.84RR295 pKa = 11.84RR296 pKa = 11.84LRR298 pKa = 11.84LRR300 pKa = 11.84VKK302 pKa = 9.95RR303 pKa = 11.84GSRR306 pKa = 11.84RR307 pKa = 11.84SLKK310 pKa = 10.31KK311 pKa = 10.36FHH313 pKa = 5.99THH315 pKa = 4.19PHH317 pKa = 5.8ARR319 pKa = 11.84QAPQAQAWSGSVVV332 pKa = 2.92
MM1 pKa = 7.1VRR3 pKa = 11.84LSRR6 pKa = 11.84LTRR9 pKa = 11.84SRR11 pKa = 11.84RR12 pKa = 11.84SLKK15 pKa = 10.24KK16 pKa = 10.48FRR18 pKa = 11.84THH20 pKa = 4.99PHH22 pKa = 5.91ARR24 pKa = 11.84QAPQAQAWSAGRR36 pKa = 11.84SQSKK40 pKa = 10.44PNGLEE45 pKa = 3.92KK46 pKa = 10.8QKK48 pKa = 9.96TNPTLRR54 pKa = 11.84GSPSRR59 pKa = 11.84RR60 pKa = 11.84LRR62 pKa = 11.84LRR64 pKa = 11.84VKK66 pKa = 10.34RR67 pKa = 11.84GSLRR71 pKa = 11.84SLKK74 pKa = 10.32KK75 pKa = 10.54FRR77 pKa = 11.84THH79 pKa = 4.99PHH81 pKa = 5.91ARR83 pKa = 11.84QAPQAQAWSAGRR95 pKa = 11.84SQSKK99 pKa = 10.44PNGLEE104 pKa = 3.92KK105 pKa = 10.8QKK107 pKa = 9.96TNPTLRR113 pKa = 11.84GSPRR117 pKa = 11.84RR118 pKa = 11.84RR119 pKa = 11.84LRR121 pKa = 11.84LRR123 pKa = 11.84VKK125 pKa = 9.95RR126 pKa = 11.84GSRR129 pKa = 11.84RR130 pKa = 11.84SLKK133 pKa = 10.27KK134 pKa = 10.52FRR136 pKa = 11.84THH138 pKa = 4.99PHH140 pKa = 5.91ARR142 pKa = 11.84QAPQAQAWSAGRR154 pKa = 11.84SQSKK158 pKa = 10.44PNGLEE163 pKa = 3.92KK164 pKa = 10.8QKK166 pKa = 9.96TNPTLRR172 pKa = 11.84GSPRR176 pKa = 11.84RR177 pKa = 11.84RR178 pKa = 11.84LRR180 pKa = 11.84LRR182 pKa = 11.84VKK184 pKa = 9.95RR185 pKa = 11.84GSRR188 pKa = 11.84RR189 pKa = 11.84SLKK192 pKa = 10.27KK193 pKa = 10.52FRR195 pKa = 11.84THH197 pKa = 4.99PHH199 pKa = 5.91ARR201 pKa = 11.84QAPQAQAWSAGRR213 pKa = 11.84SQSKK217 pKa = 10.44PNGLEE222 pKa = 3.92KK223 pKa = 10.8QKK225 pKa = 9.96TNPTLRR231 pKa = 11.84GSPRR235 pKa = 11.84RR236 pKa = 11.84RR237 pKa = 11.84LRR239 pKa = 11.84LRR241 pKa = 11.84VKK243 pKa = 9.93RR244 pKa = 11.84DD245 pKa = 2.99SRR247 pKa = 11.84RR248 pKa = 11.84SLKK251 pKa = 10.4KK252 pKa = 10.21FHH254 pKa = 5.99THH256 pKa = 4.19PHH258 pKa = 5.8ARR260 pKa = 11.84QAPQAQAWSAGRR272 pKa = 11.84SQSKK276 pKa = 10.44PNGLEE281 pKa = 3.92KK282 pKa = 10.8QKK284 pKa = 9.96TNPTLRR290 pKa = 11.84GSPRR294 pKa = 11.84RR295 pKa = 11.84RR296 pKa = 11.84LRR298 pKa = 11.84LRR300 pKa = 11.84VKK302 pKa = 9.95RR303 pKa = 11.84GSRR306 pKa = 11.84RR307 pKa = 11.84SLKK310 pKa = 10.31KK311 pKa = 10.36FHH313 pKa = 5.99THH315 pKa = 4.19PHH317 pKa = 5.8ARR319 pKa = 11.84QAPQAQAWSGSVVV332 pKa = 2.92
Molecular weight: 38.25 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2373878 |
29 |
9958 |
410.8 |
45.1 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.552 ± 0.04 | 1.1 ± 0.015 |
6.352 ± 0.042 | 5.85 ± 0.034 |
3.482 ± 0.02 | 7.657 ± 0.046 |
2.235 ± 0.019 | 4.541 ± 0.019 |
2.999 ± 0.033 | 10.052 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.147 ± 0.019 | 2.962 ± 0.032 |
5.522 ± 0.023 | 4.772 ± 0.029 |
6.866 ± 0.05 | 6.19 ± 0.036 |
5.599 ± 0.046 | 7.213 ± 0.028 |
1.519 ± 0.015 | 2.389 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
