
Ateline herpesvirus 3 (AtHV-3) (Herpesvirus ateles)
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Peploviricota; Herviviricetes; Herpesvirales; Herpesviridae; Gammaherpesvirinae; Rhadinovirus
Average proteome isoelectric point is 6.9
Get precalculated fractions of proteins
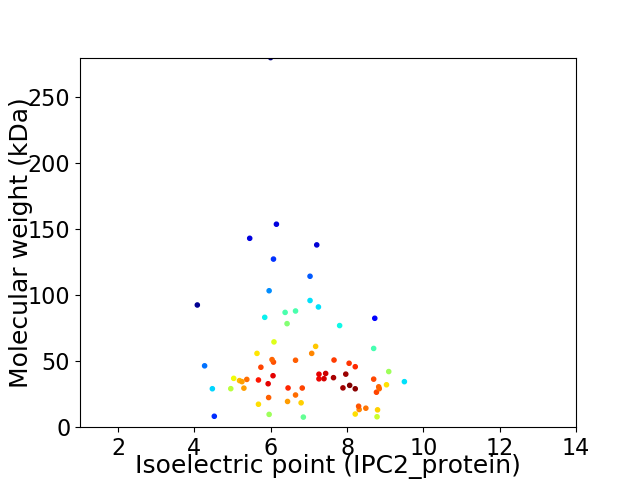
Virtual 2D-PAGE plot for 71 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q9YTL8|Q9YTL8_ATHV3 Envelope glycoprotein L OS=Ateline herpesvirus 3 OX=85618 GN=gL PE=3 SV=1
MM1 pKa = 7.3EE2 pKa = 5.72LSVPVAGVTQSNKK15 pKa = 10.18EE16 pKa = 3.65EE17 pKa = 4.23WNNIWRR23 pKa = 11.84NFEE26 pKa = 3.67QMPNVSRR33 pKa = 11.84TLSLLRR39 pKa = 11.84RR40 pKa = 11.84LFFKK44 pKa = 10.85SDD46 pKa = 3.17LGFLSSVVILKK57 pKa = 10.34QYY59 pKa = 10.62VEE61 pKa = 4.14NLEE64 pKa = 4.16TTKK67 pKa = 10.82QKK69 pKa = 11.06LSLIEE74 pKa = 3.91CLQGIKK80 pKa = 10.65YY81 pKa = 10.15LIHH84 pKa = 6.74SLYY87 pKa = 11.08KK88 pKa = 10.48NIKK91 pKa = 8.04DD92 pKa = 3.5QCDD95 pKa = 3.29VTLSINEE102 pKa = 3.91MFYY105 pKa = 11.01DD106 pKa = 4.09CKK108 pKa = 10.68TRR110 pKa = 11.84LSLILEE116 pKa = 4.4EE117 pKa = 4.39KK118 pKa = 9.9CGCGACCATSAALSKK133 pKa = 10.07IGHH136 pKa = 6.97RR137 pKa = 11.84GRR139 pKa = 11.84PPKK142 pKa = 9.53LTPHH146 pKa = 7.21KK147 pKa = 9.7SNCSAQSVLTSVHH160 pKa = 4.59NHH162 pKa = 6.64IILGINPGISEE173 pKa = 4.06WMVPEE178 pKa = 4.74IMFLPSDD185 pKa = 3.49YY186 pKa = 10.94HH187 pKa = 7.44DD188 pKa = 4.13FSEE191 pKa = 4.21HH192 pKa = 6.63KK193 pKa = 10.33KK194 pKa = 10.62DD195 pKa = 3.08ISLIATCLNCCWLYY209 pKa = 11.16FMTYY213 pKa = 9.53QYY215 pKa = 10.51MSSDD219 pKa = 4.32LIAIEE224 pKa = 4.34EE225 pKa = 4.26ALQSSYY231 pKa = 11.61LSICGSTYY239 pKa = 10.22PSYY242 pKa = 11.58SKK244 pKa = 10.52ILEE247 pKa = 4.39LLTANMSKK255 pKa = 9.67EE256 pKa = 4.18HH257 pKa = 6.29IRR259 pKa = 11.84QKK261 pKa = 11.25VNVTDD266 pKa = 4.98FIKK269 pKa = 10.67PSLHH273 pKa = 6.43QMIRR277 pKa = 11.84DD278 pKa = 3.81TKK280 pKa = 10.58KK281 pKa = 10.39EE282 pKa = 3.86PRR284 pKa = 11.84QKK286 pKa = 9.76TKK288 pKa = 10.0TLMISILGSRR298 pKa = 11.84GIGLDD303 pKa = 3.46LFRR306 pKa = 11.84TQDD309 pKa = 3.41VLKK312 pKa = 10.61FPSSDD317 pKa = 3.1AKK319 pKa = 10.82FMAVSQPDD327 pKa = 3.42NFNEE331 pKa = 4.19KK332 pKa = 9.86EE333 pKa = 4.28VEE335 pKa = 4.0FSMTGGKK342 pKa = 8.07TDD344 pKa = 3.93SEE346 pKa = 4.78DD347 pKa = 3.06VTAPRR352 pKa = 11.84KK353 pKa = 9.36VGKK356 pKa = 10.16NSLNRR361 pKa = 11.84KK362 pKa = 7.91YY363 pKa = 11.05LEE365 pKa = 3.87NLKK368 pKa = 10.79DD369 pKa = 3.78NKK371 pKa = 10.32RR372 pKa = 11.84KK373 pKa = 9.74NNNYY377 pKa = 9.73SGRR380 pKa = 11.84NNKK383 pKa = 9.69YY384 pKa = 10.41KK385 pKa = 10.92GDD387 pKa = 3.78GANDD391 pKa = 3.48KK392 pKa = 10.98DD393 pKa = 4.25KK394 pKa = 11.47SIDD397 pKa = 3.54KK398 pKa = 10.82NEE400 pKa = 4.22SEE402 pKa = 4.78GGDD405 pKa = 3.03HH406 pKa = 7.43SEE408 pKa = 4.39INRR411 pKa = 11.84EE412 pKa = 3.97KK413 pKa = 10.39NRR415 pKa = 11.84KK416 pKa = 8.02RR417 pKa = 11.84KK418 pKa = 9.43KK419 pKa = 9.19PNGFRR424 pKa = 11.84VGDD427 pKa = 3.78KK428 pKa = 10.68EE429 pKa = 4.16VGEE432 pKa = 4.26EE433 pKa = 4.03KK434 pKa = 10.33SVKK437 pKa = 10.29SGEE440 pKa = 4.37GKK442 pKa = 10.43KK443 pKa = 9.99SEE445 pKa = 4.15KK446 pKa = 10.47DD447 pKa = 3.51SEE449 pKa = 4.29EE450 pKa = 3.86EE451 pKa = 4.81AEE453 pKa = 5.36DD454 pKa = 4.61KK455 pKa = 11.37DD456 pKa = 4.18EE457 pKa = 4.83EE458 pKa = 4.28EE459 pKa = 4.31NKK461 pKa = 10.59KK462 pKa = 10.85KK463 pKa = 10.88GDD465 pKa = 3.77GEE467 pKa = 3.93EE468 pKa = 4.42DD469 pKa = 3.59EE470 pKa = 4.76EE471 pKa = 5.38DD472 pKa = 3.66EE473 pKa = 4.37EE474 pKa = 5.1EE475 pKa = 4.07EE476 pKa = 4.51DD477 pKa = 3.76EE478 pKa = 4.37EE479 pKa = 4.5EE480 pKa = 4.27EE481 pKa = 4.19EE482 pKa = 4.31EE483 pKa = 4.28EE484 pKa = 4.34EE485 pKa = 4.22EE486 pKa = 4.62EE487 pKa = 3.98EE488 pKa = 4.79DD489 pKa = 3.76EE490 pKa = 4.39EE491 pKa = 4.68EE492 pKa = 4.3EE493 pKa = 4.36EE494 pKa = 4.3EE495 pKa = 4.84EE496 pKa = 4.03EE497 pKa = 4.81DD498 pKa = 4.54EE499 pKa = 4.15EE500 pKa = 5.57DD501 pKa = 3.77EE502 pKa = 4.69EE503 pKa = 4.65EE504 pKa = 4.78EE505 pKa = 3.98EE506 pKa = 4.99DD507 pKa = 4.88EE508 pKa = 4.4EE509 pKa = 5.6DD510 pKa = 4.69EE511 pKa = 4.35EE512 pKa = 5.63DD513 pKa = 4.67EE514 pKa = 4.24EE515 pKa = 5.63DD516 pKa = 4.67EE517 pKa = 4.24EE518 pKa = 5.63DD519 pKa = 4.67EE520 pKa = 4.24EE521 pKa = 5.63DD522 pKa = 4.67EE523 pKa = 4.24EE524 pKa = 5.63DD525 pKa = 4.67EE526 pKa = 4.24EE527 pKa = 5.5DD528 pKa = 4.27EE529 pKa = 4.1EE530 pKa = 5.58DD531 pKa = 3.81EE532 pKa = 4.6EE533 pKa = 4.55EE534 pKa = 4.56EE535 pKa = 3.91EE536 pKa = 4.97DD537 pKa = 4.02EE538 pKa = 4.81EE539 pKa = 4.65EE540 pKa = 4.8EE541 pKa = 4.04EE542 pKa = 4.76DD543 pKa = 4.5EE544 pKa = 4.12EE545 pKa = 5.43DD546 pKa = 3.7EE547 pKa = 4.34EE548 pKa = 4.61EE549 pKa = 4.29EE550 pKa = 4.16EE551 pKa = 4.26EE552 pKa = 4.25EE553 pKa = 4.18EE554 pKa = 4.32EE555 pKa = 4.29EE556 pKa = 4.47EE557 pKa = 4.33EE558 pKa = 4.87EE559 pKa = 4.12EE560 pKa = 5.04DD561 pKa = 4.92EE562 pKa = 4.53EE563 pKa = 5.46DD564 pKa = 5.01EE565 pKa = 4.12EE566 pKa = 5.59DD567 pKa = 3.85EE568 pKa = 4.46EE569 pKa = 4.3EE570 pKa = 5.78KK571 pKa = 10.85EE572 pKa = 4.38DD573 pKa = 3.95EE574 pKa = 4.24EE575 pKa = 4.26EE576 pKa = 5.59KK577 pKa = 10.83EE578 pKa = 4.16DD579 pKa = 3.95EE580 pKa = 4.24EE581 pKa = 4.29EE582 pKa = 5.46KK583 pKa = 10.84EE584 pKa = 4.13DD585 pKa = 5.05EE586 pKa = 4.06EE587 pKa = 5.58DD588 pKa = 3.83EE589 pKa = 4.35EE590 pKa = 4.45EE591 pKa = 5.41KK592 pKa = 11.01EE593 pKa = 3.96DD594 pKa = 5.55DD595 pKa = 3.58EE596 pKa = 6.31DD597 pKa = 4.39EE598 pKa = 4.47EE599 pKa = 4.52EE600 pKa = 4.31EE601 pKa = 4.36EE602 pKa = 4.06EE603 pKa = 4.95GEE605 pKa = 4.2EE606 pKa = 5.29KK607 pKa = 10.64EE608 pKa = 5.22DD609 pKa = 5.22DD610 pKa = 3.76EE611 pKa = 7.11DD612 pKa = 5.66DD613 pKa = 3.98EE614 pKa = 4.81EE615 pKa = 5.7EE616 pKa = 4.18EE617 pKa = 5.28DD618 pKa = 4.37EE619 pKa = 4.69EE620 pKa = 6.02DD621 pKa = 4.92DD622 pKa = 4.74EE623 pKa = 4.86EE624 pKa = 5.45EE625 pKa = 4.03EE626 pKa = 4.84DD627 pKa = 4.87EE628 pKa = 4.4EE629 pKa = 5.63EE630 pKa = 4.08EE631 pKa = 5.75DD632 pKa = 4.59EE633 pKa = 5.51DD634 pKa = 5.8EE635 pKa = 4.3EE636 pKa = 6.42DD637 pKa = 4.22EE638 pKa = 5.38DD639 pKa = 5.85EE640 pKa = 4.29EE641 pKa = 6.4DD642 pKa = 4.22EE643 pKa = 5.38DD644 pKa = 5.85EE645 pKa = 4.29EE646 pKa = 6.4DD647 pKa = 3.61EE648 pKa = 5.55DD649 pKa = 5.29EE650 pKa = 4.43EE651 pKa = 5.71DD652 pKa = 4.18EE653 pKa = 4.8EE654 pKa = 5.34GDD656 pKa = 3.71RR657 pKa = 11.84DD658 pKa = 3.58RR659 pKa = 11.84DD660 pKa = 3.35ADD662 pKa = 3.68ADD664 pKa = 4.3RR665 pKa = 11.84DD666 pKa = 4.07GGDD669 pKa = 3.46GDD671 pKa = 4.44GDD673 pKa = 4.16GVGYY677 pKa = 10.3DD678 pKa = 3.78YY679 pKa = 11.46KK680 pKa = 11.15DD681 pKa = 3.53EE682 pKa = 4.39EE683 pKa = 4.88KK684 pKa = 10.73GTDD687 pKa = 3.07SYY689 pKa = 12.11KK690 pKa = 11.1NEE692 pKa = 3.75TEE694 pKa = 4.26YY695 pKa = 11.62NNKK698 pKa = 7.55EE699 pKa = 4.0QKK701 pKa = 10.46DD702 pKa = 3.99EE703 pKa = 4.13NLHH706 pKa = 6.23GEE708 pKa = 4.2SKK710 pKa = 10.67YY711 pKa = 10.63YY712 pKa = 10.59EE713 pKa = 3.96PVFFKK718 pKa = 10.7PKK720 pKa = 9.76QHH722 pKa = 6.35TEE724 pKa = 3.87NLSDD728 pKa = 3.72YY729 pKa = 10.63YY730 pKa = 11.32NWLRR734 pKa = 11.84NSMLSTSDD742 pKa = 3.12KK743 pKa = 10.97KK744 pKa = 11.2SISSEE749 pKa = 4.23DD750 pKa = 3.18ILEE753 pKa = 4.49NNSSYY758 pKa = 11.38LPIDD762 pKa = 3.52HH763 pKa = 7.05FEE765 pKa = 4.49KK766 pKa = 10.29ILYY769 pKa = 9.8IKK771 pKa = 8.72TPKK774 pKa = 9.33VVKK777 pKa = 9.93EE778 pKa = 4.13FQSPTDD784 pKa = 3.22SVYY787 pKa = 10.19FRR789 pKa = 11.84KK790 pKa = 10.36KK791 pKa = 9.43II792 pKa = 3.46
MM1 pKa = 7.3EE2 pKa = 5.72LSVPVAGVTQSNKK15 pKa = 10.18EE16 pKa = 3.65EE17 pKa = 4.23WNNIWRR23 pKa = 11.84NFEE26 pKa = 3.67QMPNVSRR33 pKa = 11.84TLSLLRR39 pKa = 11.84RR40 pKa = 11.84LFFKK44 pKa = 10.85SDD46 pKa = 3.17LGFLSSVVILKK57 pKa = 10.34QYY59 pKa = 10.62VEE61 pKa = 4.14NLEE64 pKa = 4.16TTKK67 pKa = 10.82QKK69 pKa = 11.06LSLIEE74 pKa = 3.91CLQGIKK80 pKa = 10.65YY81 pKa = 10.15LIHH84 pKa = 6.74SLYY87 pKa = 11.08KK88 pKa = 10.48NIKK91 pKa = 8.04DD92 pKa = 3.5QCDD95 pKa = 3.29VTLSINEE102 pKa = 3.91MFYY105 pKa = 11.01DD106 pKa = 4.09CKK108 pKa = 10.68TRR110 pKa = 11.84LSLILEE116 pKa = 4.4EE117 pKa = 4.39KK118 pKa = 9.9CGCGACCATSAALSKK133 pKa = 10.07IGHH136 pKa = 6.97RR137 pKa = 11.84GRR139 pKa = 11.84PPKK142 pKa = 9.53LTPHH146 pKa = 7.21KK147 pKa = 9.7SNCSAQSVLTSVHH160 pKa = 4.59NHH162 pKa = 6.64IILGINPGISEE173 pKa = 4.06WMVPEE178 pKa = 4.74IMFLPSDD185 pKa = 3.49YY186 pKa = 10.94HH187 pKa = 7.44DD188 pKa = 4.13FSEE191 pKa = 4.21HH192 pKa = 6.63KK193 pKa = 10.33KK194 pKa = 10.62DD195 pKa = 3.08ISLIATCLNCCWLYY209 pKa = 11.16FMTYY213 pKa = 9.53QYY215 pKa = 10.51MSSDD219 pKa = 4.32LIAIEE224 pKa = 4.34EE225 pKa = 4.26ALQSSYY231 pKa = 11.61LSICGSTYY239 pKa = 10.22PSYY242 pKa = 11.58SKK244 pKa = 10.52ILEE247 pKa = 4.39LLTANMSKK255 pKa = 9.67EE256 pKa = 4.18HH257 pKa = 6.29IRR259 pKa = 11.84QKK261 pKa = 11.25VNVTDD266 pKa = 4.98FIKK269 pKa = 10.67PSLHH273 pKa = 6.43QMIRR277 pKa = 11.84DD278 pKa = 3.81TKK280 pKa = 10.58KK281 pKa = 10.39EE282 pKa = 3.86PRR284 pKa = 11.84QKK286 pKa = 9.76TKK288 pKa = 10.0TLMISILGSRR298 pKa = 11.84GIGLDD303 pKa = 3.46LFRR306 pKa = 11.84TQDD309 pKa = 3.41VLKK312 pKa = 10.61FPSSDD317 pKa = 3.1AKK319 pKa = 10.82FMAVSQPDD327 pKa = 3.42NFNEE331 pKa = 4.19KK332 pKa = 9.86EE333 pKa = 4.28VEE335 pKa = 4.0FSMTGGKK342 pKa = 8.07TDD344 pKa = 3.93SEE346 pKa = 4.78DD347 pKa = 3.06VTAPRR352 pKa = 11.84KK353 pKa = 9.36VGKK356 pKa = 10.16NSLNRR361 pKa = 11.84KK362 pKa = 7.91YY363 pKa = 11.05LEE365 pKa = 3.87NLKK368 pKa = 10.79DD369 pKa = 3.78NKK371 pKa = 10.32RR372 pKa = 11.84KK373 pKa = 9.74NNNYY377 pKa = 9.73SGRR380 pKa = 11.84NNKK383 pKa = 9.69YY384 pKa = 10.41KK385 pKa = 10.92GDD387 pKa = 3.78GANDD391 pKa = 3.48KK392 pKa = 10.98DD393 pKa = 4.25KK394 pKa = 11.47SIDD397 pKa = 3.54KK398 pKa = 10.82NEE400 pKa = 4.22SEE402 pKa = 4.78GGDD405 pKa = 3.03HH406 pKa = 7.43SEE408 pKa = 4.39INRR411 pKa = 11.84EE412 pKa = 3.97KK413 pKa = 10.39NRR415 pKa = 11.84KK416 pKa = 8.02RR417 pKa = 11.84KK418 pKa = 9.43KK419 pKa = 9.19PNGFRR424 pKa = 11.84VGDD427 pKa = 3.78KK428 pKa = 10.68EE429 pKa = 4.16VGEE432 pKa = 4.26EE433 pKa = 4.03KK434 pKa = 10.33SVKK437 pKa = 10.29SGEE440 pKa = 4.37GKK442 pKa = 10.43KK443 pKa = 9.99SEE445 pKa = 4.15KK446 pKa = 10.47DD447 pKa = 3.51SEE449 pKa = 4.29EE450 pKa = 3.86EE451 pKa = 4.81AEE453 pKa = 5.36DD454 pKa = 4.61KK455 pKa = 11.37DD456 pKa = 4.18EE457 pKa = 4.83EE458 pKa = 4.28EE459 pKa = 4.31NKK461 pKa = 10.59KK462 pKa = 10.85KK463 pKa = 10.88GDD465 pKa = 3.77GEE467 pKa = 3.93EE468 pKa = 4.42DD469 pKa = 3.59EE470 pKa = 4.76EE471 pKa = 5.38DD472 pKa = 3.66EE473 pKa = 4.37EE474 pKa = 5.1EE475 pKa = 4.07EE476 pKa = 4.51DD477 pKa = 3.76EE478 pKa = 4.37EE479 pKa = 4.5EE480 pKa = 4.27EE481 pKa = 4.19EE482 pKa = 4.31EE483 pKa = 4.28EE484 pKa = 4.34EE485 pKa = 4.22EE486 pKa = 4.62EE487 pKa = 3.98EE488 pKa = 4.79DD489 pKa = 3.76EE490 pKa = 4.39EE491 pKa = 4.68EE492 pKa = 4.3EE493 pKa = 4.36EE494 pKa = 4.3EE495 pKa = 4.84EE496 pKa = 4.03EE497 pKa = 4.81DD498 pKa = 4.54EE499 pKa = 4.15EE500 pKa = 5.57DD501 pKa = 3.77EE502 pKa = 4.69EE503 pKa = 4.65EE504 pKa = 4.78EE505 pKa = 3.98EE506 pKa = 4.99DD507 pKa = 4.88EE508 pKa = 4.4EE509 pKa = 5.6DD510 pKa = 4.69EE511 pKa = 4.35EE512 pKa = 5.63DD513 pKa = 4.67EE514 pKa = 4.24EE515 pKa = 5.63DD516 pKa = 4.67EE517 pKa = 4.24EE518 pKa = 5.63DD519 pKa = 4.67EE520 pKa = 4.24EE521 pKa = 5.63DD522 pKa = 4.67EE523 pKa = 4.24EE524 pKa = 5.63DD525 pKa = 4.67EE526 pKa = 4.24EE527 pKa = 5.5DD528 pKa = 4.27EE529 pKa = 4.1EE530 pKa = 5.58DD531 pKa = 3.81EE532 pKa = 4.6EE533 pKa = 4.55EE534 pKa = 4.56EE535 pKa = 3.91EE536 pKa = 4.97DD537 pKa = 4.02EE538 pKa = 4.81EE539 pKa = 4.65EE540 pKa = 4.8EE541 pKa = 4.04EE542 pKa = 4.76DD543 pKa = 4.5EE544 pKa = 4.12EE545 pKa = 5.43DD546 pKa = 3.7EE547 pKa = 4.34EE548 pKa = 4.61EE549 pKa = 4.29EE550 pKa = 4.16EE551 pKa = 4.26EE552 pKa = 4.25EE553 pKa = 4.18EE554 pKa = 4.32EE555 pKa = 4.29EE556 pKa = 4.47EE557 pKa = 4.33EE558 pKa = 4.87EE559 pKa = 4.12EE560 pKa = 5.04DD561 pKa = 4.92EE562 pKa = 4.53EE563 pKa = 5.46DD564 pKa = 5.01EE565 pKa = 4.12EE566 pKa = 5.59DD567 pKa = 3.85EE568 pKa = 4.46EE569 pKa = 4.3EE570 pKa = 5.78KK571 pKa = 10.85EE572 pKa = 4.38DD573 pKa = 3.95EE574 pKa = 4.24EE575 pKa = 4.26EE576 pKa = 5.59KK577 pKa = 10.83EE578 pKa = 4.16DD579 pKa = 3.95EE580 pKa = 4.24EE581 pKa = 4.29EE582 pKa = 5.46KK583 pKa = 10.84EE584 pKa = 4.13DD585 pKa = 5.05EE586 pKa = 4.06EE587 pKa = 5.58DD588 pKa = 3.83EE589 pKa = 4.35EE590 pKa = 4.45EE591 pKa = 5.41KK592 pKa = 11.01EE593 pKa = 3.96DD594 pKa = 5.55DD595 pKa = 3.58EE596 pKa = 6.31DD597 pKa = 4.39EE598 pKa = 4.47EE599 pKa = 4.52EE600 pKa = 4.31EE601 pKa = 4.36EE602 pKa = 4.06EE603 pKa = 4.95GEE605 pKa = 4.2EE606 pKa = 5.29KK607 pKa = 10.64EE608 pKa = 5.22DD609 pKa = 5.22DD610 pKa = 3.76EE611 pKa = 7.11DD612 pKa = 5.66DD613 pKa = 3.98EE614 pKa = 4.81EE615 pKa = 5.7EE616 pKa = 4.18EE617 pKa = 5.28DD618 pKa = 4.37EE619 pKa = 4.69EE620 pKa = 6.02DD621 pKa = 4.92DD622 pKa = 4.74EE623 pKa = 4.86EE624 pKa = 5.45EE625 pKa = 4.03EE626 pKa = 4.84DD627 pKa = 4.87EE628 pKa = 4.4EE629 pKa = 5.63EE630 pKa = 4.08EE631 pKa = 5.75DD632 pKa = 4.59EE633 pKa = 5.51DD634 pKa = 5.8EE635 pKa = 4.3EE636 pKa = 6.42DD637 pKa = 4.22EE638 pKa = 5.38DD639 pKa = 5.85EE640 pKa = 4.29EE641 pKa = 6.4DD642 pKa = 4.22EE643 pKa = 5.38DD644 pKa = 5.85EE645 pKa = 4.29EE646 pKa = 6.4DD647 pKa = 3.61EE648 pKa = 5.55DD649 pKa = 5.29EE650 pKa = 4.43EE651 pKa = 5.71DD652 pKa = 4.18EE653 pKa = 4.8EE654 pKa = 5.34GDD656 pKa = 3.71RR657 pKa = 11.84DD658 pKa = 3.58RR659 pKa = 11.84DD660 pKa = 3.35ADD662 pKa = 3.68ADD664 pKa = 4.3RR665 pKa = 11.84DD666 pKa = 4.07GGDD669 pKa = 3.46GDD671 pKa = 4.44GDD673 pKa = 4.16GVGYY677 pKa = 10.3DD678 pKa = 3.78YY679 pKa = 11.46KK680 pKa = 11.15DD681 pKa = 3.53EE682 pKa = 4.39EE683 pKa = 4.88KK684 pKa = 10.73GTDD687 pKa = 3.07SYY689 pKa = 12.11KK690 pKa = 11.1NEE692 pKa = 3.75TEE694 pKa = 4.26YY695 pKa = 11.62NNKK698 pKa = 7.55EE699 pKa = 4.0QKK701 pKa = 10.46DD702 pKa = 3.99EE703 pKa = 4.13NLHH706 pKa = 6.23GEE708 pKa = 4.2SKK710 pKa = 10.67YY711 pKa = 10.63YY712 pKa = 10.59EE713 pKa = 3.96PVFFKK718 pKa = 10.7PKK720 pKa = 9.76QHH722 pKa = 6.35TEE724 pKa = 3.87NLSDD728 pKa = 3.72YY729 pKa = 10.63YY730 pKa = 11.32NWLRR734 pKa = 11.84NSMLSTSDD742 pKa = 3.12KK743 pKa = 10.97KK744 pKa = 11.2SISSEE749 pKa = 4.23DD750 pKa = 3.18ILEE753 pKa = 4.49NNSSYY758 pKa = 11.38LPIDD762 pKa = 3.52HH763 pKa = 7.05FEE765 pKa = 4.49KK766 pKa = 10.29ILYY769 pKa = 9.8IKK771 pKa = 8.72TPKK774 pKa = 9.33VVKK777 pKa = 9.93EE778 pKa = 4.13FQSPTDD784 pKa = 3.22SVYY787 pKa = 10.19FRR789 pKa = 11.84KK790 pKa = 10.36KK791 pKa = 9.43II792 pKa = 3.46
Molecular weight: 92.59 kDa
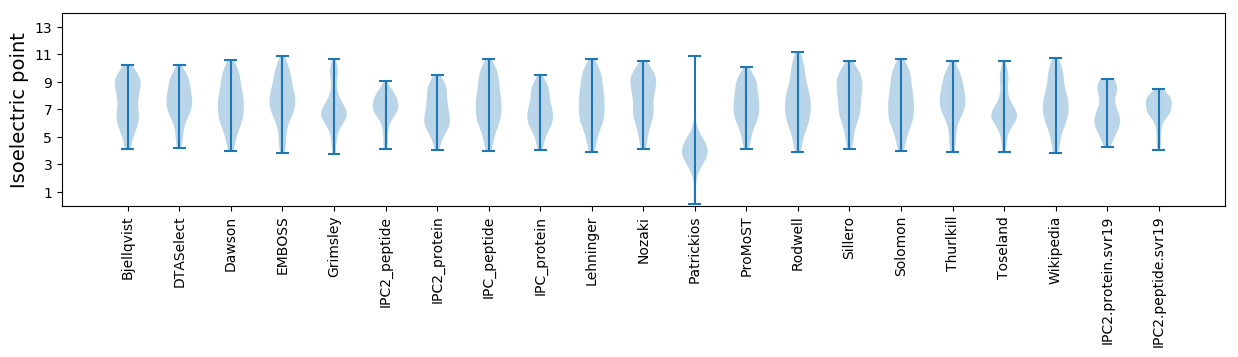
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q9YTP7|Q9YTP7_ATHV3 Capsid vertex component 2 OS=Ateline herpesvirus 3 OX=85618 GN=CVC2 PE=3 SV=1
MM1 pKa = 7.5LSVIQKK7 pKa = 7.44TNKK10 pKa = 9.71DD11 pKa = 3.36ALAQLPKK18 pKa = 9.25TRR20 pKa = 11.84KK21 pKa = 8.32IAGNKK26 pKa = 8.72AHH28 pKa = 7.24IATYY32 pKa = 10.3QKK34 pKa = 10.34LIKK37 pKa = 10.4YY38 pKa = 7.43SAPTPLFKK46 pKa = 10.7FLGISHH52 pKa = 7.48PCAVQTKK59 pKa = 8.34NQLFFEE65 pKa = 4.42VRR67 pKa = 11.84LGNRR71 pKa = 11.84IADD74 pKa = 4.26CIMLTSCGEE83 pKa = 3.8TRR85 pKa = 11.84ICYY88 pKa = 9.91VIEE91 pKa = 5.04LKK93 pKa = 9.7TCMTSSLDD101 pKa = 3.47LSSDD105 pKa = 2.53IRR107 pKa = 11.84QTQRR111 pKa = 11.84SQGLCQLADD120 pKa = 3.51TVSFIQNYY128 pKa = 9.35APPGRR133 pKa = 11.84QAWTVLPILIFKK145 pKa = 10.14SQATLKK151 pKa = 8.47TLHH154 pKa = 6.62VEE156 pKa = 4.05IPKK159 pKa = 10.11FPANLTHH166 pKa = 6.52TSEE169 pKa = 4.47EE170 pKa = 4.07KK171 pKa = 10.48LSCFFFSRR179 pKa = 11.84SDD181 pKa = 3.33VEE183 pKa = 4.01IRR185 pKa = 11.84KK186 pKa = 9.36KK187 pKa = 10.88LHH189 pKa = 6.09FCSKK193 pKa = 9.7TKK195 pKa = 10.63TNTQWDD201 pKa = 4.18SVLDD205 pKa = 3.81PTSTKK210 pKa = 10.31QSVYY214 pKa = 9.09RR215 pKa = 11.84QRR217 pKa = 11.84IIDD220 pKa = 3.83RR221 pKa = 11.84NKK223 pKa = 9.78KK224 pKa = 9.27KK225 pKa = 10.57CFSIQVSSFKK235 pKa = 10.66CRR237 pKa = 11.84NRR239 pKa = 11.84PNKK242 pKa = 9.83KK243 pKa = 9.66INGKK247 pKa = 9.36LRR249 pKa = 11.84TGQADD254 pKa = 3.2ARR256 pKa = 11.84PFEE259 pKa = 4.65KK260 pKa = 10.24KK261 pKa = 7.01QHH263 pKa = 4.76NNKK266 pKa = 9.9RR267 pKa = 11.84LGNNRR272 pKa = 11.84KK273 pKa = 9.28HH274 pKa = 5.87GRR276 pKa = 11.84QATRR280 pKa = 11.84YY281 pKa = 7.79QANAPLFSATLISNNPAHH299 pKa = 6.52ALKK302 pKa = 10.62RR303 pKa = 3.82
MM1 pKa = 7.5LSVIQKK7 pKa = 7.44TNKK10 pKa = 9.71DD11 pKa = 3.36ALAQLPKK18 pKa = 9.25TRR20 pKa = 11.84KK21 pKa = 8.32IAGNKK26 pKa = 8.72AHH28 pKa = 7.24IATYY32 pKa = 10.3QKK34 pKa = 10.34LIKK37 pKa = 10.4YY38 pKa = 7.43SAPTPLFKK46 pKa = 10.7FLGISHH52 pKa = 7.48PCAVQTKK59 pKa = 8.34NQLFFEE65 pKa = 4.42VRR67 pKa = 11.84LGNRR71 pKa = 11.84IADD74 pKa = 4.26CIMLTSCGEE83 pKa = 3.8TRR85 pKa = 11.84ICYY88 pKa = 9.91VIEE91 pKa = 5.04LKK93 pKa = 9.7TCMTSSLDD101 pKa = 3.47LSSDD105 pKa = 2.53IRR107 pKa = 11.84QTQRR111 pKa = 11.84SQGLCQLADD120 pKa = 3.51TVSFIQNYY128 pKa = 9.35APPGRR133 pKa = 11.84QAWTVLPILIFKK145 pKa = 10.14SQATLKK151 pKa = 8.47TLHH154 pKa = 6.62VEE156 pKa = 4.05IPKK159 pKa = 10.11FPANLTHH166 pKa = 6.52TSEE169 pKa = 4.47EE170 pKa = 4.07KK171 pKa = 10.48LSCFFFSRR179 pKa = 11.84SDD181 pKa = 3.33VEE183 pKa = 4.01IRR185 pKa = 11.84KK186 pKa = 9.36KK187 pKa = 10.88LHH189 pKa = 6.09FCSKK193 pKa = 9.7TKK195 pKa = 10.63TNTQWDD201 pKa = 4.18SVLDD205 pKa = 3.81PTSTKK210 pKa = 10.31QSVYY214 pKa = 9.09RR215 pKa = 11.84QRR217 pKa = 11.84IIDD220 pKa = 3.83RR221 pKa = 11.84NKK223 pKa = 9.78KK224 pKa = 9.27KK225 pKa = 10.57CFSIQVSSFKK235 pKa = 10.66CRR237 pKa = 11.84NRR239 pKa = 11.84PNKK242 pKa = 9.83KK243 pKa = 9.66INGKK247 pKa = 9.36LRR249 pKa = 11.84TGQADD254 pKa = 3.2ARR256 pKa = 11.84PFEE259 pKa = 4.65KK260 pKa = 10.24KK261 pKa = 7.01QHH263 pKa = 4.76NNKK266 pKa = 9.9RR267 pKa = 11.84LGNNRR272 pKa = 11.84KK273 pKa = 9.28HH274 pKa = 5.87GRR276 pKa = 11.84QATRR280 pKa = 11.84YY281 pKa = 7.79QANAPLFSATLISNNPAHH299 pKa = 6.52ALKK302 pKa = 10.62RR303 pKa = 3.82
Molecular weight: 34.5 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
32476 |
66 |
2471 |
457.4 |
51.78 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.994 ± 0.241 | 2.525 ± 0.184 |
4.582 ± 0.259 | 5.724 ± 0.466 |
4.859 ± 0.185 | 4.329 ± 0.3 |
2.694 ± 0.087 | 7.439 ± 0.272 |
6.719 ± 0.264 | 10.112 ± 0.217 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.242 ± 0.104 | 5.632 ± 0.174 |
5.099 ± 0.267 | 3.904 ± 0.189 |
3.218 ± 0.165 | 8.166 ± 0.205 |
7.045 ± 0.298 | 5.86 ± 0.26 |
1.075 ± 0.08 | 3.781 ± 0.157 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
