
Beihai sobemo-like virus 12
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.1
Get precalculated fractions of proteins
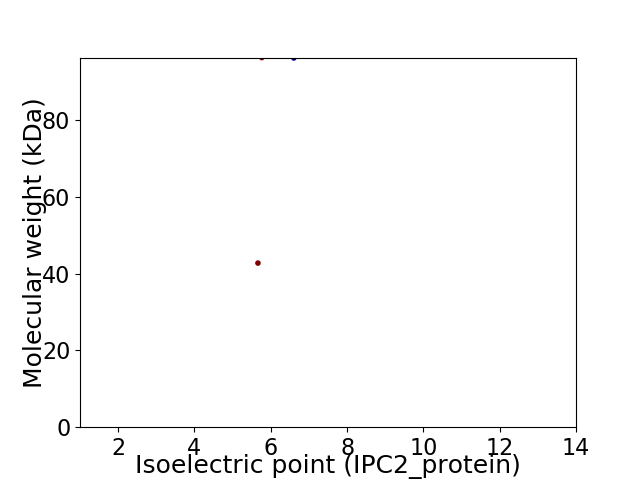
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KED7|A0A1L3KED7_9VIRU RNA-directed RNA polymerase OS=Beihai sobemo-like virus 12 OX=1922683 PE=4 SV=1
MM1 pKa = 7.99LEE3 pKa = 4.34DD4 pKa = 3.49VTTFTSWINNINRR17 pKa = 11.84EE18 pKa = 4.24TMRR21 pKa = 11.84MDD23 pKa = 4.68PDD25 pKa = 3.83SSPGLPWGGTNGDD38 pKa = 3.32RR39 pKa = 11.84FGYY42 pKa = 9.16VPCAGYY48 pKa = 10.55DD49 pKa = 3.63PNKK52 pKa = 10.36LDD54 pKa = 4.14EE55 pKa = 4.76LRR57 pKa = 11.84MAVLIRR63 pKa = 11.84IEE65 pKa = 4.05EE66 pKa = 4.3LKK68 pKa = 10.67VAPALDD74 pKa = 4.6PIFVFIKK81 pKa = 10.62QEE83 pKa = 3.64MHH85 pKa = 6.51KK86 pKa = 10.29KK87 pKa = 9.43SKK89 pKa = 10.42AVEE92 pKa = 4.13GRR94 pKa = 11.84WRR96 pKa = 11.84LICGVGLTDD105 pKa = 4.3QIVARR110 pKa = 11.84ILFEE114 pKa = 4.51PYY116 pKa = 10.02FDD118 pKa = 4.84FLLDD122 pKa = 3.49HH123 pKa = 6.53PLIYY127 pKa = 9.33KK128 pKa = 7.74TAIGWGLTSHH138 pKa = 6.93GSLSYY143 pKa = 10.86MNFFLGGGQLQAADD157 pKa = 3.6KK158 pKa = 10.86SAWDD162 pKa = 3.43WTVQPWVFEE171 pKa = 4.13VFKK174 pKa = 10.86EE175 pKa = 4.27IMHH178 pKa = 6.28KK179 pKa = 8.42MHH181 pKa = 7.52IGRR184 pKa = 11.84EE185 pKa = 4.3DD186 pKa = 3.16LWHH189 pKa = 6.37QLMEE193 pKa = 3.93HH194 pKa = 6.87HH195 pKa = 7.15IDD197 pKa = 4.23AICYY201 pKa = 9.8HH202 pKa = 6.6KK203 pKa = 11.12VLDD206 pKa = 3.67IVGRR210 pKa = 11.84KK211 pKa = 9.03LDD213 pKa = 4.3CPPGIMPSGWFMTIAFNSVAQLILHH238 pKa = 7.31AYY240 pKa = 10.04ADD242 pKa = 4.53DD243 pKa = 3.93NNEE246 pKa = 3.85FDD248 pKa = 3.73WPFVMGDD255 pKa = 3.43DD256 pKa = 5.06TIQKK260 pKa = 8.91PASDD264 pKa = 4.4RR265 pKa = 11.84YY266 pKa = 7.25WKK268 pKa = 10.84RR269 pKa = 11.84MTTTGAKK276 pKa = 9.77IKK278 pKa = 10.59DD279 pKa = 3.21IVTTRR284 pKa = 11.84EE285 pKa = 3.52FVGFVFKK292 pKa = 10.78DD293 pKa = 3.53RR294 pKa = 11.84SYY296 pKa = 10.95HH297 pKa = 5.51PSYY300 pKa = 9.09TVKK303 pKa = 10.8YY304 pKa = 7.82NCKK307 pKa = 9.12MNNIPPEE314 pKa = 4.14VLVEE318 pKa = 4.17TLTSYY323 pKa = 9.78QWLYY327 pKa = 11.28AFEE330 pKa = 4.34PEE332 pKa = 4.05KK333 pKa = 11.03LKK335 pKa = 10.64IIHH338 pKa = 7.21RR339 pKa = 11.84YY340 pKa = 6.0MCEE343 pKa = 3.49IGASSHH349 pKa = 5.93IVTSSEE355 pKa = 3.63MADD358 pKa = 2.88RR359 pKa = 11.84VLGLKK364 pKa = 9.74SLPVGSSWW372 pKa = 3.74
MM1 pKa = 7.99LEE3 pKa = 4.34DD4 pKa = 3.49VTTFTSWINNINRR17 pKa = 11.84EE18 pKa = 4.24TMRR21 pKa = 11.84MDD23 pKa = 4.68PDD25 pKa = 3.83SSPGLPWGGTNGDD38 pKa = 3.32RR39 pKa = 11.84FGYY42 pKa = 9.16VPCAGYY48 pKa = 10.55DD49 pKa = 3.63PNKK52 pKa = 10.36LDD54 pKa = 4.14EE55 pKa = 4.76LRR57 pKa = 11.84MAVLIRR63 pKa = 11.84IEE65 pKa = 4.05EE66 pKa = 4.3LKK68 pKa = 10.67VAPALDD74 pKa = 4.6PIFVFIKK81 pKa = 10.62QEE83 pKa = 3.64MHH85 pKa = 6.51KK86 pKa = 10.29KK87 pKa = 9.43SKK89 pKa = 10.42AVEE92 pKa = 4.13GRR94 pKa = 11.84WRR96 pKa = 11.84LICGVGLTDD105 pKa = 4.3QIVARR110 pKa = 11.84ILFEE114 pKa = 4.51PYY116 pKa = 10.02FDD118 pKa = 4.84FLLDD122 pKa = 3.49HH123 pKa = 6.53PLIYY127 pKa = 9.33KK128 pKa = 7.74TAIGWGLTSHH138 pKa = 6.93GSLSYY143 pKa = 10.86MNFFLGGGQLQAADD157 pKa = 3.6KK158 pKa = 10.86SAWDD162 pKa = 3.43WTVQPWVFEE171 pKa = 4.13VFKK174 pKa = 10.86EE175 pKa = 4.27IMHH178 pKa = 6.28KK179 pKa = 8.42MHH181 pKa = 7.52IGRR184 pKa = 11.84EE185 pKa = 4.3DD186 pKa = 3.16LWHH189 pKa = 6.37QLMEE193 pKa = 3.93HH194 pKa = 6.87HH195 pKa = 7.15IDD197 pKa = 4.23AICYY201 pKa = 9.8HH202 pKa = 6.6KK203 pKa = 11.12VLDD206 pKa = 3.67IVGRR210 pKa = 11.84KK211 pKa = 9.03LDD213 pKa = 4.3CPPGIMPSGWFMTIAFNSVAQLILHH238 pKa = 7.31AYY240 pKa = 10.04ADD242 pKa = 4.53DD243 pKa = 3.93NNEE246 pKa = 3.85FDD248 pKa = 3.73WPFVMGDD255 pKa = 3.43DD256 pKa = 5.06TIQKK260 pKa = 8.91PASDD264 pKa = 4.4RR265 pKa = 11.84YY266 pKa = 7.25WKK268 pKa = 10.84RR269 pKa = 11.84MTTTGAKK276 pKa = 9.77IKK278 pKa = 10.59DD279 pKa = 3.21IVTTRR284 pKa = 11.84EE285 pKa = 3.52FVGFVFKK292 pKa = 10.78DD293 pKa = 3.53RR294 pKa = 11.84SYY296 pKa = 10.95HH297 pKa = 5.51PSYY300 pKa = 9.09TVKK303 pKa = 10.8YY304 pKa = 7.82NCKK307 pKa = 9.12MNNIPPEE314 pKa = 4.14VLVEE318 pKa = 4.17TLTSYY323 pKa = 9.78QWLYY327 pKa = 11.28AFEE330 pKa = 4.34PEE332 pKa = 4.05KK333 pKa = 11.03LKK335 pKa = 10.64IIHH338 pKa = 7.21RR339 pKa = 11.84YY340 pKa = 6.0MCEE343 pKa = 3.49IGASSHH349 pKa = 5.93IVTSSEE355 pKa = 3.63MADD358 pKa = 2.88RR359 pKa = 11.84VLGLKK364 pKa = 9.74SLPVGSSWW372 pKa = 3.74
Molecular weight: 42.79 kDa
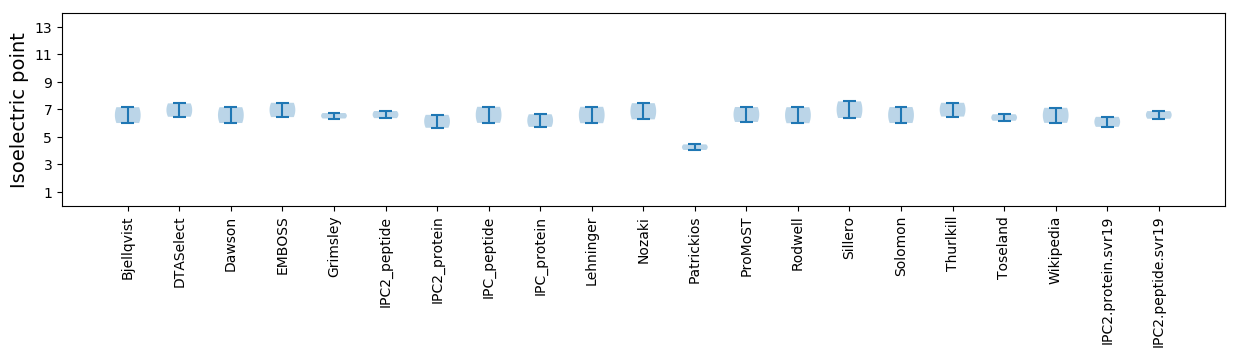
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KED7|A0A1L3KED7_9VIRU RNA-directed RNA polymerase OS=Beihai sobemo-like virus 12 OX=1922683 PE=4 SV=1
MM1 pKa = 7.85AAQQVDD7 pKa = 3.88TFFQQWMQTSWKK19 pKa = 10.42VLEE22 pKa = 3.86QDD24 pKa = 2.86VWSSYY29 pKa = 10.41IIYY32 pKa = 10.29LRR34 pKa = 11.84VVYY37 pKa = 10.41SSSSFLMDD45 pKa = 3.65VIANFCHH52 pKa = 6.25YY53 pKa = 10.65LGDD56 pKa = 3.87RR57 pKa = 11.84PGIMNFLGFLLAVLIYY73 pKa = 10.62LVLNLPIRR81 pKa = 11.84KK82 pKa = 8.86NLSSLKK88 pKa = 10.22NRR90 pKa = 11.84YY91 pKa = 8.72KK92 pKa = 11.0DD93 pKa = 3.3SFLGSWICFFLSGFIMITKK112 pKa = 9.48YY113 pKa = 10.89AVISLLVMILWLLLTRR129 pKa = 11.84IILTTALLRR138 pKa = 11.84TGFPRR143 pKa = 11.84AVVDD147 pKa = 4.51FVVGAYY153 pKa = 9.83AAGDD157 pKa = 3.67VIVTYY162 pKa = 10.8KK163 pKa = 10.9FITLFVLGYY172 pKa = 9.39KK173 pKa = 8.78VARR176 pKa = 11.84ASFEE180 pKa = 4.24VKK182 pKa = 10.0IQADD186 pKa = 3.84EE187 pKa = 4.14SDD189 pKa = 4.08SIKK192 pKa = 10.62IPGTTEE198 pKa = 3.17IDD200 pKa = 3.53MNSCKK205 pKa = 10.43SAVAVGTIKK214 pKa = 10.64NGKK217 pKa = 9.07FNIVGQATCINAHH230 pKa = 6.33CLGIDD235 pKa = 4.04GVADD239 pKa = 3.69TFLTATHH246 pKa = 6.49IVKK249 pKa = 10.47AFPIDD254 pKa = 4.45DD255 pKa = 4.23LYY257 pKa = 10.83IWRR260 pKa = 11.84NGVAVKK266 pKa = 10.65VGLGLQCDD274 pKa = 3.84PDD276 pKa = 4.17GLFDD280 pKa = 4.06IYY282 pKa = 10.86LLRR285 pKa = 11.84VRR287 pKa = 11.84DD288 pKa = 3.9GASKK292 pKa = 10.83LGVSPAKK299 pKa = 10.18VANFPLGKK307 pKa = 10.07CAGTLFSISTEE318 pKa = 3.89GLLQSVGMIKK328 pKa = 10.02IATDD332 pKa = 3.54QEE334 pKa = 4.13DD335 pKa = 3.53SNYY338 pKa = 9.77TFIGNTIEE346 pKa = 4.16GWSGSGLYY354 pKa = 10.53VSGALVGMHH363 pKa = 5.87TVRR366 pKa = 11.84LGTDD370 pKa = 2.73KK371 pKa = 10.99KK372 pKa = 11.21LNAGYY377 pKa = 10.26DD378 pKa = 3.76ISWVPHH384 pKa = 5.74LVRR387 pKa = 11.84STLPLQHH394 pKa = 6.89RR395 pKa = 11.84CKK397 pKa = 10.21QCNEE401 pKa = 4.42EE402 pKa = 3.79IHH404 pKa = 6.85HH405 pKa = 6.43FDD407 pKa = 2.95ISEE410 pKa = 4.28GFDD413 pKa = 3.04IPKK416 pKa = 10.09KK417 pKa = 9.35QGKK420 pKa = 9.25KK421 pKa = 10.54GEE423 pKa = 4.5EE424 pKa = 3.75ICLSCDD430 pKa = 3.2NEE432 pKa = 4.4NEE434 pKa = 3.68HH435 pKa = 7.0SYY437 pKa = 11.0PFEE440 pKa = 4.48KK441 pKa = 10.15KK442 pKa = 10.24GKK444 pKa = 8.32NKK446 pKa = 10.45SSISDD451 pKa = 2.89TWDD454 pKa = 3.18YY455 pKa = 11.72LDD457 pKa = 4.66RR458 pKa = 11.84QLKK461 pKa = 9.48QDD463 pKa = 3.35STKK466 pKa = 10.65VKK468 pKa = 7.66KK469 pKa = 8.98TKK471 pKa = 9.31WGLEE475 pKa = 3.61NLYY478 pKa = 10.71AITLEE483 pKa = 3.89KK484 pKa = 10.61GKK486 pKa = 10.63VYY488 pKa = 10.8RR489 pKa = 11.84FTDD492 pKa = 4.06DD493 pKa = 4.51EE494 pKa = 5.95LDD496 pKa = 3.68TQLGAEE502 pKa = 4.07RR503 pKa = 11.84AKK505 pKa = 10.69VIRR508 pKa = 11.84ARR510 pKa = 11.84AEE512 pKa = 4.52GYY514 pKa = 10.13SSNSRR519 pKa = 11.84SDD521 pKa = 3.4YY522 pKa = 10.41FGEE525 pKa = 4.46SKK527 pKa = 11.12FEE529 pKa = 4.28GKK531 pKa = 10.58GPAQIDD537 pKa = 3.76SKK539 pKa = 10.63RR540 pKa = 11.84RR541 pKa = 11.84PIAPVDD547 pKa = 3.67LHH549 pKa = 6.59HH550 pKa = 7.02LQLARR555 pKa = 11.84LWIQNIEE562 pKa = 4.1NTQSQVSGLIRR573 pKa = 11.84ALSQLMITEE582 pKa = 4.04RR583 pKa = 11.84RR584 pKa = 11.84NWEE587 pKa = 3.74ASEE590 pKa = 4.39SEE592 pKa = 3.86EE593 pKa = 4.92DD594 pKa = 3.2MGKK597 pKa = 9.99IIQTQVDD604 pKa = 3.55IRR606 pKa = 11.84SLQLFLEE613 pKa = 4.67QIAEE617 pKa = 4.35YY618 pKa = 10.52NFEE621 pKa = 4.07ATEE624 pKa = 4.64LYY626 pKa = 10.35QLVEE630 pKa = 3.95IGSNVDD636 pKa = 2.68ITLKK640 pKa = 10.72NRR642 pKa = 11.84VEE644 pKa = 4.12RR645 pKa = 11.84QKK647 pKa = 11.2KK648 pKa = 9.98SYY650 pKa = 10.5LKK652 pKa = 9.83EE653 pKa = 4.17HH654 pKa = 7.08PLPTTSPDD662 pKa = 2.95IDD664 pKa = 3.87LTIDD668 pKa = 3.29NADD671 pKa = 3.9RR672 pKa = 11.84NLTEE676 pKa = 4.04GLEE679 pKa = 4.04ILRR682 pKa = 11.84NIPKK686 pKa = 10.36RR687 pKa = 11.84LTPEE691 pKa = 3.69DD692 pKa = 3.73TASVEE697 pKa = 4.15AAFGPVIAEE706 pKa = 3.63ILKK709 pKa = 10.36RR710 pKa = 11.84RR711 pKa = 11.84GIVAQLPMDD720 pKa = 3.95WEE722 pKa = 4.72NNDD725 pKa = 3.23GMDD728 pKa = 5.06PEE730 pKa = 5.63FDD732 pKa = 5.4DD733 pKa = 6.33DD734 pKa = 6.57DD735 pKa = 4.63EE736 pKa = 4.83EE737 pKa = 6.0QDD739 pKa = 3.8FQDD742 pKa = 4.13PRR744 pKa = 11.84STEE747 pKa = 3.54QPAPSKK753 pKa = 10.71GKK755 pKa = 10.06KK756 pKa = 9.06AALGQTTKK764 pKa = 10.97GKK766 pKa = 5.99TTSAKK771 pKa = 10.59SPATSTKK778 pKa = 10.35QSMSQEE784 pKa = 3.88RR785 pKa = 11.84KK786 pKa = 8.92PRR788 pKa = 11.84SLSRR792 pKa = 11.84GNSQRR797 pKa = 11.84TQSMSNLRR805 pKa = 11.84KK806 pKa = 9.9LEE808 pKa = 3.91KK809 pKa = 10.29EE810 pKa = 4.12KK811 pKa = 10.66QLKK814 pKa = 8.54AQYY817 pKa = 10.49EE818 pKa = 4.64SILSYY823 pKa = 7.84TQKK826 pKa = 10.4QKK828 pKa = 11.32KK829 pKa = 8.29EE830 pKa = 3.99LEE832 pKa = 4.15SLQMEE837 pKa = 4.57ISDD840 pKa = 4.21GLLKK844 pKa = 10.62EE845 pKa = 4.07SSAYY849 pKa = 9.68FRR851 pKa = 11.84KK852 pKa = 9.37QQTKK856 pKa = 9.91
MM1 pKa = 7.85AAQQVDD7 pKa = 3.88TFFQQWMQTSWKK19 pKa = 10.42VLEE22 pKa = 3.86QDD24 pKa = 2.86VWSSYY29 pKa = 10.41IIYY32 pKa = 10.29LRR34 pKa = 11.84VVYY37 pKa = 10.41SSSSFLMDD45 pKa = 3.65VIANFCHH52 pKa = 6.25YY53 pKa = 10.65LGDD56 pKa = 3.87RR57 pKa = 11.84PGIMNFLGFLLAVLIYY73 pKa = 10.62LVLNLPIRR81 pKa = 11.84KK82 pKa = 8.86NLSSLKK88 pKa = 10.22NRR90 pKa = 11.84YY91 pKa = 8.72KK92 pKa = 11.0DD93 pKa = 3.3SFLGSWICFFLSGFIMITKK112 pKa = 9.48YY113 pKa = 10.89AVISLLVMILWLLLTRR129 pKa = 11.84IILTTALLRR138 pKa = 11.84TGFPRR143 pKa = 11.84AVVDD147 pKa = 4.51FVVGAYY153 pKa = 9.83AAGDD157 pKa = 3.67VIVTYY162 pKa = 10.8KK163 pKa = 10.9FITLFVLGYY172 pKa = 9.39KK173 pKa = 8.78VARR176 pKa = 11.84ASFEE180 pKa = 4.24VKK182 pKa = 10.0IQADD186 pKa = 3.84EE187 pKa = 4.14SDD189 pKa = 4.08SIKK192 pKa = 10.62IPGTTEE198 pKa = 3.17IDD200 pKa = 3.53MNSCKK205 pKa = 10.43SAVAVGTIKK214 pKa = 10.64NGKK217 pKa = 9.07FNIVGQATCINAHH230 pKa = 6.33CLGIDD235 pKa = 4.04GVADD239 pKa = 3.69TFLTATHH246 pKa = 6.49IVKK249 pKa = 10.47AFPIDD254 pKa = 4.45DD255 pKa = 4.23LYY257 pKa = 10.83IWRR260 pKa = 11.84NGVAVKK266 pKa = 10.65VGLGLQCDD274 pKa = 3.84PDD276 pKa = 4.17GLFDD280 pKa = 4.06IYY282 pKa = 10.86LLRR285 pKa = 11.84VRR287 pKa = 11.84DD288 pKa = 3.9GASKK292 pKa = 10.83LGVSPAKK299 pKa = 10.18VANFPLGKK307 pKa = 10.07CAGTLFSISTEE318 pKa = 3.89GLLQSVGMIKK328 pKa = 10.02IATDD332 pKa = 3.54QEE334 pKa = 4.13DD335 pKa = 3.53SNYY338 pKa = 9.77TFIGNTIEE346 pKa = 4.16GWSGSGLYY354 pKa = 10.53VSGALVGMHH363 pKa = 5.87TVRR366 pKa = 11.84LGTDD370 pKa = 2.73KK371 pKa = 10.99KK372 pKa = 11.21LNAGYY377 pKa = 10.26DD378 pKa = 3.76ISWVPHH384 pKa = 5.74LVRR387 pKa = 11.84STLPLQHH394 pKa = 6.89RR395 pKa = 11.84CKK397 pKa = 10.21QCNEE401 pKa = 4.42EE402 pKa = 3.79IHH404 pKa = 6.85HH405 pKa = 6.43FDD407 pKa = 2.95ISEE410 pKa = 4.28GFDD413 pKa = 3.04IPKK416 pKa = 10.09KK417 pKa = 9.35QGKK420 pKa = 9.25KK421 pKa = 10.54GEE423 pKa = 4.5EE424 pKa = 3.75ICLSCDD430 pKa = 3.2NEE432 pKa = 4.4NEE434 pKa = 3.68HH435 pKa = 7.0SYY437 pKa = 11.0PFEE440 pKa = 4.48KK441 pKa = 10.15KK442 pKa = 10.24GKK444 pKa = 8.32NKK446 pKa = 10.45SSISDD451 pKa = 2.89TWDD454 pKa = 3.18YY455 pKa = 11.72LDD457 pKa = 4.66RR458 pKa = 11.84QLKK461 pKa = 9.48QDD463 pKa = 3.35STKK466 pKa = 10.65VKK468 pKa = 7.66KK469 pKa = 8.98TKK471 pKa = 9.31WGLEE475 pKa = 3.61NLYY478 pKa = 10.71AITLEE483 pKa = 3.89KK484 pKa = 10.61GKK486 pKa = 10.63VYY488 pKa = 10.8RR489 pKa = 11.84FTDD492 pKa = 4.06DD493 pKa = 4.51EE494 pKa = 5.95LDD496 pKa = 3.68TQLGAEE502 pKa = 4.07RR503 pKa = 11.84AKK505 pKa = 10.69VIRR508 pKa = 11.84ARR510 pKa = 11.84AEE512 pKa = 4.52GYY514 pKa = 10.13SSNSRR519 pKa = 11.84SDD521 pKa = 3.4YY522 pKa = 10.41FGEE525 pKa = 4.46SKK527 pKa = 11.12FEE529 pKa = 4.28GKK531 pKa = 10.58GPAQIDD537 pKa = 3.76SKK539 pKa = 10.63RR540 pKa = 11.84RR541 pKa = 11.84PIAPVDD547 pKa = 3.67LHH549 pKa = 6.59HH550 pKa = 7.02LQLARR555 pKa = 11.84LWIQNIEE562 pKa = 4.1NTQSQVSGLIRR573 pKa = 11.84ALSQLMITEE582 pKa = 4.04RR583 pKa = 11.84RR584 pKa = 11.84NWEE587 pKa = 3.74ASEE590 pKa = 4.39SEE592 pKa = 3.86EE593 pKa = 4.92DD594 pKa = 3.2MGKK597 pKa = 9.99IIQTQVDD604 pKa = 3.55IRR606 pKa = 11.84SLQLFLEE613 pKa = 4.67QIAEE617 pKa = 4.35YY618 pKa = 10.52NFEE621 pKa = 4.07ATEE624 pKa = 4.64LYY626 pKa = 10.35QLVEE630 pKa = 3.95IGSNVDD636 pKa = 2.68ITLKK640 pKa = 10.72NRR642 pKa = 11.84VEE644 pKa = 4.12RR645 pKa = 11.84QKK647 pKa = 11.2KK648 pKa = 9.98SYY650 pKa = 10.5LKK652 pKa = 9.83EE653 pKa = 4.17HH654 pKa = 7.08PLPTTSPDD662 pKa = 2.95IDD664 pKa = 3.87LTIDD668 pKa = 3.29NADD671 pKa = 3.9RR672 pKa = 11.84NLTEE676 pKa = 4.04GLEE679 pKa = 4.04ILRR682 pKa = 11.84NIPKK686 pKa = 10.36RR687 pKa = 11.84LTPEE691 pKa = 3.69DD692 pKa = 3.73TASVEE697 pKa = 4.15AAFGPVIAEE706 pKa = 3.63ILKK709 pKa = 10.36RR710 pKa = 11.84RR711 pKa = 11.84GIVAQLPMDD720 pKa = 3.95WEE722 pKa = 4.72NNDD725 pKa = 3.23GMDD728 pKa = 5.06PEE730 pKa = 5.63FDD732 pKa = 5.4DD733 pKa = 6.33DD734 pKa = 6.57DD735 pKa = 4.63EE736 pKa = 4.83EE737 pKa = 6.0QDD739 pKa = 3.8FQDD742 pKa = 4.13PRR744 pKa = 11.84STEE747 pKa = 3.54QPAPSKK753 pKa = 10.71GKK755 pKa = 10.06KK756 pKa = 9.06AALGQTTKK764 pKa = 10.97GKK766 pKa = 5.99TTSAKK771 pKa = 10.59SPATSTKK778 pKa = 10.35QSMSQEE784 pKa = 3.88RR785 pKa = 11.84KK786 pKa = 8.92PRR788 pKa = 11.84SLSRR792 pKa = 11.84GNSQRR797 pKa = 11.84TQSMSNLRR805 pKa = 11.84KK806 pKa = 9.9LEE808 pKa = 3.91KK809 pKa = 10.29EE810 pKa = 4.12KK811 pKa = 10.66QLKK814 pKa = 8.54AQYY817 pKa = 10.49EE818 pKa = 4.64SILSYY823 pKa = 7.84TQKK826 pKa = 10.4QKK828 pKa = 11.32KK829 pKa = 8.29EE830 pKa = 3.99LEE832 pKa = 4.15SLQMEE837 pKa = 4.57ISDD840 pKa = 4.21GLLKK844 pKa = 10.62EE845 pKa = 4.07SSAYY849 pKa = 9.68FRR851 pKa = 11.84KK852 pKa = 9.37QQTKK856 pKa = 9.91
Molecular weight: 96.26 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1228 |
372 |
856 |
614.0 |
69.53 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.945 ± 0.295 | 1.384 ± 0.119 |
6.352 ± 0.192 | 5.945 ± 0.295 |
4.316 ± 0.411 | 6.596 ± 0.065 |
2.036 ± 0.758 | 7.166 ± 0.048 |
7.085 ± 0.608 | 9.446 ± 0.718 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.606 ± 0.881 | 3.664 ± 0.228 |
3.909 ± 0.763 | 4.397 ± 1.028 |
4.642 ± 0.177 | 7.41 ± 0.917 |
5.782 ± 0.211 | 5.863 ± 0.306 |
2.117 ± 0.716 | 3.339 ± 0.221 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
