
Enterovibrio sp. CAIM 600
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Vibrionales; Vibrionaceae; Enterovibrio; unclassified Enterovibrio
Average proteome isoelectric point is 6.14
Get precalculated fractions of proteins
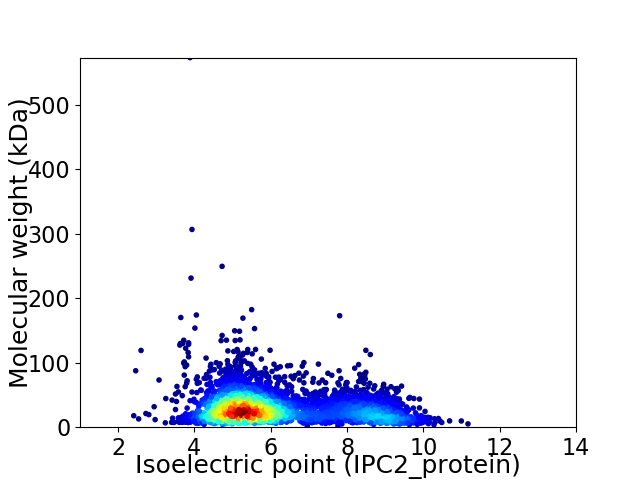
Virtual 2D-PAGE plot for 4013 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4Q0YMC7|A0A4Q0YMC7_9GAMM Thiol-disulfide oxidoreductase OS=Enterovibrio sp. CAIM 600 OX=1278244 GN=CS022_20475 PE=4 SV=1
MM1 pKa = 7.6VYY3 pKa = 9.59TLNRR7 pKa = 11.84DD8 pKa = 3.23DD9 pKa = 5.25SGYY12 pKa = 8.89LTASDD17 pKa = 4.23TVTDD21 pKa = 4.09TLTLTATDD29 pKa = 4.07GTQQQITISITGTDD43 pKa = 3.19TTAIISGNKK52 pKa = 9.13QSTLLKK58 pKa = 10.14TEE60 pKa = 4.17TTASGTVSLFDD71 pKa = 5.15PDD73 pKa = 5.51ASTQPTLPNQTHH85 pKa = 6.77IGTYY89 pKa = 8.58GTLTTNSDD97 pKa = 3.75GTWSYY102 pKa = 11.7VVDD105 pKa = 4.54PSAIKK110 pKa = 10.49ALNDD114 pKa = 3.81DD115 pKa = 4.56EE116 pKa = 4.44QTQDD120 pKa = 3.23TFAVTASDD128 pKa = 3.67GSEE131 pKa = 3.69HH132 pKa = 6.63QIIITVTGDD141 pKa = 2.84EE142 pKa = 4.33DD143 pKa = 4.57ASVISGVTSGSVAEE157 pKa = 5.08DD158 pKa = 3.73DD159 pKa = 4.83ADD161 pKa = 5.97LIAQGQLSITDD172 pKa = 3.75EE173 pKa = 4.39DD174 pKa = 4.79TNDD177 pKa = 3.59SPSLINTTLTGQYY190 pKa = 9.1GTFDD194 pKa = 4.52LNNGHH199 pKa = 5.85WTFTANNSALQPLDD213 pKa = 3.39QGQTAQDD220 pKa = 3.64TFTVNASDD228 pKa = 4.11GNSEE232 pKa = 4.53LVTITLTGVEE242 pKa = 4.57DD243 pKa = 3.88ASSVGGTHH251 pKa = 6.51TGSVTEE257 pKa = 4.14NSLGPFSISAGALATGATTTSNTSSHH283 pKa = 5.38IQIAGVPYY291 pKa = 10.44ILTTNVGYY299 pKa = 10.76NGLVTFHH306 pKa = 6.97RR307 pKa = 11.84VEE309 pKa = 3.96NDD311 pKa = 3.15GSVTEE316 pKa = 4.24SDD318 pKa = 3.2RR319 pKa = 11.84MFYY322 pKa = 10.64DD323 pKa = 3.82SSQGTVATTLSGSITSDD340 pKa = 3.31VQSFGLPLDD349 pKa = 3.85ALGNGLTASNIYY361 pKa = 10.51DD362 pKa = 3.43VTGQTTLFLTSQNSGSISVWHH383 pKa = 6.81ISDD386 pKa = 3.24TGEE389 pKa = 4.07LSFHH393 pKa = 6.25GGKK396 pKa = 7.97TFGHH400 pKa = 5.88SQPSTNGGIVRR411 pKa = 11.84EE412 pKa = 4.27NILYY416 pKa = 7.5EE417 pKa = 4.26APGGTFYY424 pKa = 10.54IYY426 pKa = 10.05ATRR429 pKa = 11.84PQNDD433 pKa = 4.14RR434 pKa = 11.84IDD436 pKa = 3.65VLTYY440 pKa = 10.95NPTTGAISEE449 pKa = 4.41TGTTISAGDD458 pKa = 3.75NVSSLDD464 pKa = 3.87LLTHH468 pKa = 6.94GGNTYY473 pKa = 10.71LFSASNDD480 pKa = 3.54TLNVYY485 pKa = 10.59AIDD488 pKa = 3.99EE489 pKa = 4.79SNGGLTLEE497 pKa = 4.43TSNTFDD503 pKa = 3.66LGSNHH508 pKa = 6.24SVNTHH513 pKa = 3.73QTEE516 pKa = 4.23NGDD519 pKa = 3.98SFILASGDD527 pKa = 3.63DD528 pKa = 3.81NKK530 pKa = 10.46ISHH533 pKa = 7.6LYY535 pKa = 9.33QLEE538 pKa = 4.04NDD540 pKa = 3.82GSITQRR546 pKa = 11.84DD547 pKa = 3.92SLDD550 pKa = 3.32SGFSYY555 pKa = 10.42HH556 pKa = 7.34SSPTEE561 pKa = 3.73VDD563 pKa = 2.65NHH565 pKa = 6.84LIFVCEE571 pKa = 4.2SEE573 pKa = 4.3SGGVDD578 pKa = 3.95LFTIGANHH586 pKa = 7.31RR587 pKa = 11.84FQHH590 pKa = 6.03LKK592 pKa = 10.25HH593 pKa = 6.45LSGFDD598 pKa = 3.33NDD600 pKa = 4.32DD601 pKa = 3.7AVPQLVVTEE610 pKa = 4.32NGNFYY615 pKa = 10.73LVDD618 pKa = 3.3INGVFTSQALTITDD632 pKa = 4.59LTPQATGTLTISDD645 pKa = 4.47PDD647 pKa = 4.09ANDD650 pKa = 3.19TPTFQNTNVDD660 pKa = 3.34GTYY663 pKa = 11.07GSLSLTDD670 pKa = 3.69GTWFIAQQHH679 pKa = 5.73IALRR683 pKa = 11.84PP684 pKa = 3.36
MM1 pKa = 7.6VYY3 pKa = 9.59TLNRR7 pKa = 11.84DD8 pKa = 3.23DD9 pKa = 5.25SGYY12 pKa = 8.89LTASDD17 pKa = 4.23TVTDD21 pKa = 4.09TLTLTATDD29 pKa = 4.07GTQQQITISITGTDD43 pKa = 3.19TTAIISGNKK52 pKa = 9.13QSTLLKK58 pKa = 10.14TEE60 pKa = 4.17TTASGTVSLFDD71 pKa = 5.15PDD73 pKa = 5.51ASTQPTLPNQTHH85 pKa = 6.77IGTYY89 pKa = 8.58GTLTTNSDD97 pKa = 3.75GTWSYY102 pKa = 11.7VVDD105 pKa = 4.54PSAIKK110 pKa = 10.49ALNDD114 pKa = 3.81DD115 pKa = 4.56EE116 pKa = 4.44QTQDD120 pKa = 3.23TFAVTASDD128 pKa = 3.67GSEE131 pKa = 3.69HH132 pKa = 6.63QIIITVTGDD141 pKa = 2.84EE142 pKa = 4.33DD143 pKa = 4.57ASVISGVTSGSVAEE157 pKa = 5.08DD158 pKa = 3.73DD159 pKa = 4.83ADD161 pKa = 5.97LIAQGQLSITDD172 pKa = 3.75EE173 pKa = 4.39DD174 pKa = 4.79TNDD177 pKa = 3.59SPSLINTTLTGQYY190 pKa = 9.1GTFDD194 pKa = 4.52LNNGHH199 pKa = 5.85WTFTANNSALQPLDD213 pKa = 3.39QGQTAQDD220 pKa = 3.64TFTVNASDD228 pKa = 4.11GNSEE232 pKa = 4.53LVTITLTGVEE242 pKa = 4.57DD243 pKa = 3.88ASSVGGTHH251 pKa = 6.51TGSVTEE257 pKa = 4.14NSLGPFSISAGALATGATTTSNTSSHH283 pKa = 5.38IQIAGVPYY291 pKa = 10.44ILTTNVGYY299 pKa = 10.76NGLVTFHH306 pKa = 6.97RR307 pKa = 11.84VEE309 pKa = 3.96NDD311 pKa = 3.15GSVTEE316 pKa = 4.24SDD318 pKa = 3.2RR319 pKa = 11.84MFYY322 pKa = 10.64DD323 pKa = 3.82SSQGTVATTLSGSITSDD340 pKa = 3.31VQSFGLPLDD349 pKa = 3.85ALGNGLTASNIYY361 pKa = 10.51DD362 pKa = 3.43VTGQTTLFLTSQNSGSISVWHH383 pKa = 6.81ISDD386 pKa = 3.24TGEE389 pKa = 4.07LSFHH393 pKa = 6.25GGKK396 pKa = 7.97TFGHH400 pKa = 5.88SQPSTNGGIVRR411 pKa = 11.84EE412 pKa = 4.27NILYY416 pKa = 7.5EE417 pKa = 4.26APGGTFYY424 pKa = 10.54IYY426 pKa = 10.05ATRR429 pKa = 11.84PQNDD433 pKa = 4.14RR434 pKa = 11.84IDD436 pKa = 3.65VLTYY440 pKa = 10.95NPTTGAISEE449 pKa = 4.41TGTTISAGDD458 pKa = 3.75NVSSLDD464 pKa = 3.87LLTHH468 pKa = 6.94GGNTYY473 pKa = 10.71LFSASNDD480 pKa = 3.54TLNVYY485 pKa = 10.59AIDD488 pKa = 3.99EE489 pKa = 4.79SNGGLTLEE497 pKa = 4.43TSNTFDD503 pKa = 3.66LGSNHH508 pKa = 6.24SVNTHH513 pKa = 3.73QTEE516 pKa = 4.23NGDD519 pKa = 3.98SFILASGDD527 pKa = 3.63DD528 pKa = 3.81NKK530 pKa = 10.46ISHH533 pKa = 7.6LYY535 pKa = 9.33QLEE538 pKa = 4.04NDD540 pKa = 3.82GSITQRR546 pKa = 11.84DD547 pKa = 3.92SLDD550 pKa = 3.32SGFSYY555 pKa = 10.42HH556 pKa = 7.34SSPTEE561 pKa = 3.73VDD563 pKa = 2.65NHH565 pKa = 6.84LIFVCEE571 pKa = 4.2SEE573 pKa = 4.3SGGVDD578 pKa = 3.95LFTIGANHH586 pKa = 7.31RR587 pKa = 11.84FQHH590 pKa = 6.03LKK592 pKa = 10.25HH593 pKa = 6.45LSGFDD598 pKa = 3.33NDD600 pKa = 4.32DD601 pKa = 3.7AVPQLVVTEE610 pKa = 4.32NGNFYY615 pKa = 10.73LVDD618 pKa = 3.3INGVFTSQALTITDD632 pKa = 4.59LTPQATGTLTISDD645 pKa = 4.47PDD647 pKa = 4.09ANDD650 pKa = 3.19TPTFQNTNVDD660 pKa = 3.34GTYY663 pKa = 11.07GSLSLTDD670 pKa = 3.69GTWFIAQQHH679 pKa = 5.73IALRR683 pKa = 11.84PP684 pKa = 3.36
Molecular weight: 72.05 kDa
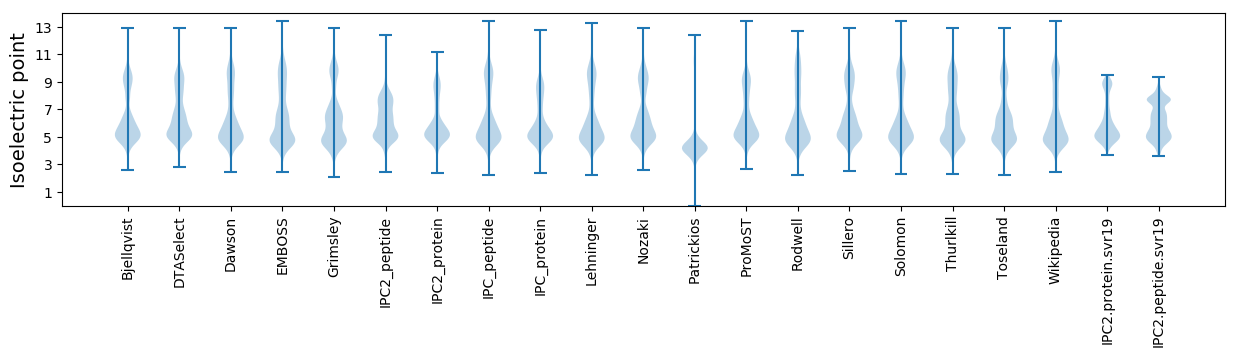
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Q0YMZ2|A0A4Q0YMZ2_9GAMM Lysine transporter LysE OS=Enterovibrio sp. CAIM 600 OX=1278244 GN=CS022_19805 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.49RR12 pKa = 11.84KK13 pKa = 7.97RR14 pKa = 11.84THH16 pKa = 5.89GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.34NGRR28 pKa = 11.84KK29 pKa = 9.74VIGARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.13GRR39 pKa = 11.84KK40 pKa = 8.23QLSKK44 pKa = 11.23
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.49RR12 pKa = 11.84KK13 pKa = 7.97RR14 pKa = 11.84THH16 pKa = 5.89GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.34NGRR28 pKa = 11.84KK29 pKa = 9.74VIGARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.13GRR39 pKa = 11.84KK40 pKa = 8.23QLSKK44 pKa = 11.23
Molecular weight: 5.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1136605 |
16 |
5376 |
283.2 |
31.46 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.235 ± 0.046 | 1.064 ± 0.017 |
5.886 ± 0.044 | 6.324 ± 0.038 |
4.287 ± 0.029 | 6.834 ± 0.036 |
2.24 ± 0.022 | 6.113 ± 0.029 |
5.325 ± 0.037 | 10.079 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.574 ± 0.023 | 4.342 ± 0.033 |
3.899 ± 0.025 | 4.067 ± 0.028 |
4.647 ± 0.037 | 7.197 ± 0.039 |
5.624 ± 0.049 | 7.039 ± 0.028 |
1.28 ± 0.016 | 2.941 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
