
Burkholderia virus phiE122
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Myoviridae; Peduovirinae; Tigrvirus
Average proteome isoelectric point is 7.0
Get precalculated fractions of proteins
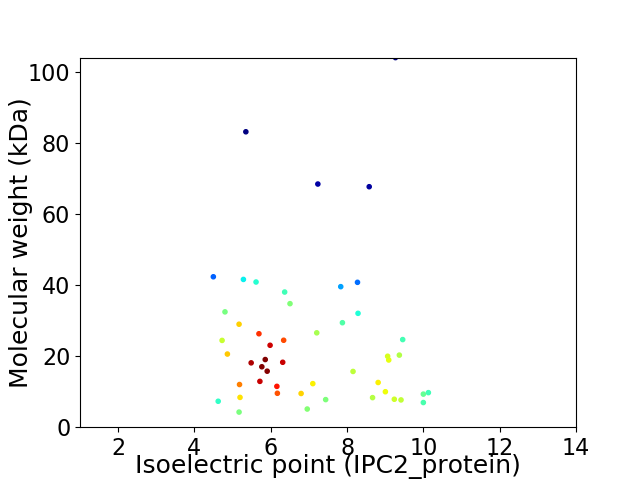
Virtual 2D-PAGE plot for 50 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A4JWW3|A4JWW3_9CAUD Uncharacterized protein OS=Burkholderia virus phiE122 OX=431892 GN=BPSphiE122_0018 PE=4 SV=1
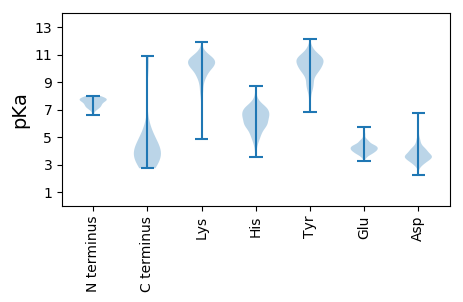
MM1 pKa = 7.8NDD3 pKa = 3.35QQQSRR8 pKa = 11.84ADD10 pKa = 3.51ALTEE14 pKa = 3.77EE15 pKa = 4.75QIRR18 pKa = 11.84AFAEE22 pKa = 3.78NAFDD26 pKa = 4.13HH27 pKa = 6.42EE28 pKa = 4.83VGAGALRR35 pKa = 11.84QNVATVGNVILAIRR49 pKa = 11.84NALNLAASANEE60 pKa = 4.05TGAEE64 pKa = 4.08GAAITDD70 pKa = 3.65HH71 pKa = 6.86DD72 pKa = 4.63AKK74 pKa = 10.7GLEE77 pKa = 4.77IIAGWLYY84 pKa = 11.23KK85 pKa = 10.68DD86 pKa = 3.98GLVEE90 pKa = 3.81PAAMLRR96 pKa = 11.84RR97 pKa = 11.84LASARR102 pKa = 11.84SPAMAAAAPADD113 pKa = 3.54EE114 pKa = 5.29HH115 pKa = 6.64STIEE119 pKa = 4.39CQAHH123 pKa = 6.14SGPDD127 pKa = 3.36CTEE130 pKa = 4.19CGGTGAWSGSADD142 pKa = 3.73EE143 pKa = 4.85RR144 pKa = 11.84AALQWAAGTLQEE156 pKa = 4.49IVSGRR161 pKa = 11.84WKK163 pKa = 10.4GAKK166 pKa = 9.84EE167 pKa = 3.82SDD169 pKa = 3.44KK170 pKa = 11.27VSIGSVTKK178 pKa = 9.9TVAQVLDD185 pKa = 3.71MADD188 pKa = 3.25AALGRR193 pKa = 11.84ASQGDD198 pKa = 3.77APAEE202 pKa = 3.93MRR204 pKa = 11.84KK205 pKa = 9.31PDD207 pKa = 4.01AYY209 pKa = 11.38VPIHH213 pKa = 6.52PRR215 pKa = 11.84NGPLWASAVTSLDD228 pKa = 2.92ADD230 pKa = 3.96RR231 pKa = 11.84PKK233 pKa = 10.62SYY235 pKa = 10.23PVQAVYY241 pKa = 10.64LGASTAALSAARR253 pKa = 11.84AAASPAADD261 pKa = 3.62EE262 pKa = 4.54AQALLTVGRR271 pKa = 11.84CMGIEE276 pKa = 3.84AVEE279 pKa = 4.32KK280 pKa = 10.72YY281 pKa = 9.91IDD283 pKa = 3.81SASLGTVIDD292 pKa = 3.84YY293 pKa = 10.47AAEE296 pKa = 3.97IRR298 pKa = 11.84KK299 pKa = 8.73LAAPQPAPADD309 pKa = 3.5APADD313 pKa = 3.76VTLIPYY319 pKa = 10.23DD320 pKa = 3.55GLTEE324 pKa = 4.05EE325 pKa = 4.83FTDD328 pKa = 4.29EE329 pKa = 4.17VARR332 pKa = 11.84LANDD336 pKa = 3.24APGIRR341 pKa = 11.84EE342 pKa = 4.06AVAGALQSCGAIIAPADD359 pKa = 3.89APAHH363 pKa = 5.83AAEE366 pKa = 5.29CPHH369 pKa = 6.95CDD371 pKa = 3.29GEE373 pKa = 4.8GVIEE377 pKa = 4.5ADD379 pKa = 4.51SGASPCACAQDD390 pKa = 3.95AQEE393 pKa = 4.3GMPTFGARR401 pKa = 11.84RR402 pKa = 11.84TQADD406 pKa = 3.57APAEE410 pKa = 4.02ARR412 pKa = 11.84VV413 pKa = 3.29
MM1 pKa = 7.8NDD3 pKa = 3.35QQQSRR8 pKa = 11.84ADD10 pKa = 3.51ALTEE14 pKa = 3.77EE15 pKa = 4.75QIRR18 pKa = 11.84AFAEE22 pKa = 3.78NAFDD26 pKa = 4.13HH27 pKa = 6.42EE28 pKa = 4.83VGAGALRR35 pKa = 11.84QNVATVGNVILAIRR49 pKa = 11.84NALNLAASANEE60 pKa = 4.05TGAEE64 pKa = 4.08GAAITDD70 pKa = 3.65HH71 pKa = 6.86DD72 pKa = 4.63AKK74 pKa = 10.7GLEE77 pKa = 4.77IIAGWLYY84 pKa = 11.23KK85 pKa = 10.68DD86 pKa = 3.98GLVEE90 pKa = 3.81PAAMLRR96 pKa = 11.84RR97 pKa = 11.84LASARR102 pKa = 11.84SPAMAAAAPADD113 pKa = 3.54EE114 pKa = 5.29HH115 pKa = 6.64STIEE119 pKa = 4.39CQAHH123 pKa = 6.14SGPDD127 pKa = 3.36CTEE130 pKa = 4.19CGGTGAWSGSADD142 pKa = 3.73EE143 pKa = 4.85RR144 pKa = 11.84AALQWAAGTLQEE156 pKa = 4.49IVSGRR161 pKa = 11.84WKK163 pKa = 10.4GAKK166 pKa = 9.84EE167 pKa = 3.82SDD169 pKa = 3.44KK170 pKa = 11.27VSIGSVTKK178 pKa = 9.9TVAQVLDD185 pKa = 3.71MADD188 pKa = 3.25AALGRR193 pKa = 11.84ASQGDD198 pKa = 3.77APAEE202 pKa = 3.93MRR204 pKa = 11.84KK205 pKa = 9.31PDD207 pKa = 4.01AYY209 pKa = 11.38VPIHH213 pKa = 6.52PRR215 pKa = 11.84NGPLWASAVTSLDD228 pKa = 2.92ADD230 pKa = 3.96RR231 pKa = 11.84PKK233 pKa = 10.62SYY235 pKa = 10.23PVQAVYY241 pKa = 10.64LGASTAALSAARR253 pKa = 11.84AAASPAADD261 pKa = 3.62EE262 pKa = 4.54AQALLTVGRR271 pKa = 11.84CMGIEE276 pKa = 3.84AVEE279 pKa = 4.32KK280 pKa = 10.72YY281 pKa = 9.91IDD283 pKa = 3.81SASLGTVIDD292 pKa = 3.84YY293 pKa = 10.47AAEE296 pKa = 3.97IRR298 pKa = 11.84KK299 pKa = 8.73LAAPQPAPADD309 pKa = 3.5APADD313 pKa = 3.76VTLIPYY319 pKa = 10.23DD320 pKa = 3.55GLTEE324 pKa = 4.05EE325 pKa = 4.83FTDD328 pKa = 4.29EE329 pKa = 4.17VARR332 pKa = 11.84LANDD336 pKa = 3.24APGIRR341 pKa = 11.84EE342 pKa = 4.06AVAGALQSCGAIIAPADD359 pKa = 3.89APAHH363 pKa = 5.83AAEE366 pKa = 5.29CPHH369 pKa = 6.95CDD371 pKa = 3.29GEE373 pKa = 4.8GVIEE377 pKa = 4.5ADD379 pKa = 4.51SGASPCACAQDD390 pKa = 3.95AQEE393 pKa = 4.3GMPTFGARR401 pKa = 11.84RR402 pKa = 11.84TQADD406 pKa = 3.57APAEE410 pKa = 4.02ARR412 pKa = 11.84VV413 pKa = 3.29
Molecular weight: 42.31 kDa
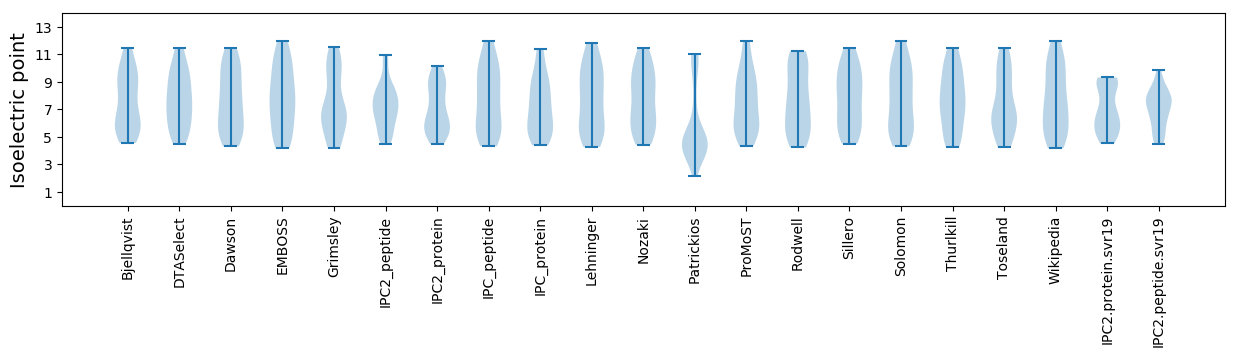
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A4JWW6|A4JWW6_9CAUD Uncharacterized protein OS=Burkholderia virus phiE122 OX=431892 GN=BPSphiE122_0021 PE=4 SV=1
MM1 pKa = 6.97EE2 pKa = 4.27QSNEE6 pKa = 3.74RR7 pKa = 11.84GVAEE11 pKa = 3.74RR12 pKa = 11.84RR13 pKa = 11.84EE14 pKa = 4.06ALFEE18 pKa = 4.51DD19 pKa = 4.45LAEE22 pKa = 4.49LGIGAGVCVPAFVVYY37 pKa = 8.84TPPRR41 pKa = 11.84TARR44 pKa = 11.84GDD46 pKa = 3.35RR47 pKa = 11.84AAANAMKK54 pKa = 10.32EE55 pKa = 4.07LRR57 pKa = 11.84PLLAKK62 pKa = 10.36LARR65 pKa = 11.84RR66 pKa = 11.84RR67 pKa = 11.84LILRR71 pKa = 3.9
MM1 pKa = 6.97EE2 pKa = 4.27QSNEE6 pKa = 3.74RR7 pKa = 11.84GVAEE11 pKa = 3.74RR12 pKa = 11.84RR13 pKa = 11.84EE14 pKa = 4.06ALFEE18 pKa = 4.51DD19 pKa = 4.45LAEE22 pKa = 4.49LGIGAGVCVPAFVVYY37 pKa = 8.84TPPRR41 pKa = 11.84TARR44 pKa = 11.84GDD46 pKa = 3.35RR47 pKa = 11.84AAANAMKK54 pKa = 10.32EE55 pKa = 4.07LRR57 pKa = 11.84PLLAKK62 pKa = 10.36LARR65 pKa = 11.84RR66 pKa = 11.84RR67 pKa = 11.84LILRR71 pKa = 3.9
Molecular weight: 7.86 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
11296 |
37 |
988 |
225.9 |
24.59 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.191 ± 0.631 | 1.027 ± 0.167 |
6.294 ± 0.343 | 5.48 ± 0.254 |
3.214 ± 0.206 | 7.994 ± 0.489 |
2.125 ± 0.199 | 4.258 ± 0.203 |
3.798 ± 0.296 | 8.401 ± 0.419 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.302 ± 0.165 | 2.797 ± 0.194 |
4.869 ± 0.34 | 3.435 ± 0.196 |
8.136 ± 0.424 | 5.046 ± 0.243 |
5.675 ± 0.282 | 7.047 ± 0.292 |
1.514 ± 0.137 | 2.399 ± 0.162 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
