
Human papillomavirus type 50
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Gammapapillomavirus; Gammapapillomavirus 3
Average proteome isoelectric point is 5.95
Get precalculated fractions of proteins
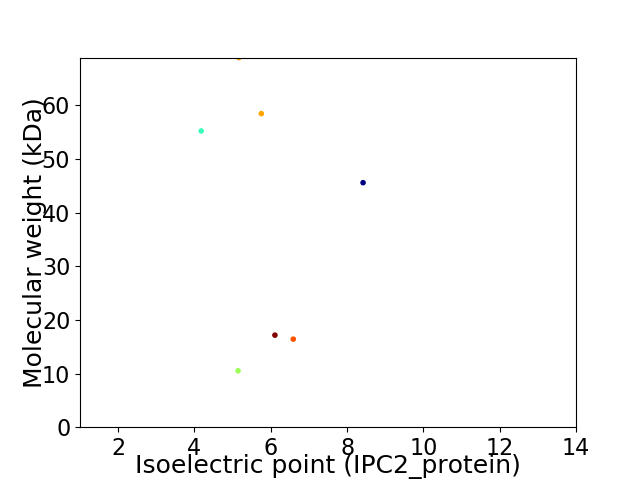
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

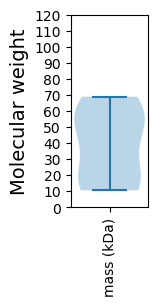
Protein with the lowest isoelectric point:
>sp|Q80932|VL2_HPV50 Minor capsid protein L2 OS=Human papillomavirus type 50 OX=40539 GN=L2 PE=3 SV=1
MM1 pKa = 7.5LRR3 pKa = 11.84RR4 pKa = 11.84RR5 pKa = 11.84KK6 pKa = 9.54RR7 pKa = 11.84ASPTDD12 pKa = 4.02LYY14 pKa = 10.94RR15 pKa = 11.84SCLQGGDD22 pKa = 4.9CIPDD26 pKa = 3.37VQNKK30 pKa = 9.65FEE32 pKa = 4.96GNTIADD38 pKa = 3.83WLLKK42 pKa = 10.38IFGGLVYY49 pKa = 10.47FGNLGIGTGRR59 pKa = 11.84GTGGTFGYY67 pKa = 10.22RR68 pKa = 11.84PFGAPGSGRR77 pKa = 11.84PTQEE81 pKa = 4.06LPIARR86 pKa = 11.84PNVVIDD92 pKa = 4.1PLGPAPIVPVDD103 pKa = 3.96PSAASIVPLVEE114 pKa = 4.01GAPDD118 pKa = 3.39VGFAAPDD125 pKa = 3.7AGPAAGGTDD134 pKa = 2.95IEE136 pKa = 5.77LYY138 pKa = 10.36TITNSTTDD146 pKa = 3.08VGAVGGGPTVTSNEE160 pKa = 4.03EE161 pKa = 3.81FEE163 pKa = 4.65VAVIDD168 pKa = 4.12AQPIAPYY175 pKa = 9.75PKK177 pKa = 9.61QLLYY181 pKa = 11.04DD182 pKa = 3.6STIAATFEE190 pKa = 4.2TQINPFINPDD200 pKa = 3.17INNVNVLVDD209 pKa = 3.59PSFAGDD215 pKa = 3.44TVGDD219 pKa = 3.75YY220 pKa = 10.51FYY222 pKa = 11.5EE223 pKa = 4.62EE224 pKa = 4.36IPLEE228 pKa = 4.14RR229 pKa = 11.84LDD231 pKa = 3.68IQTFDD236 pKa = 3.1ILEE239 pKa = 4.72PPTEE243 pKa = 4.22STPTQLGNRR252 pKa = 11.84FVSRR256 pKa = 11.84ARR258 pKa = 11.84DD259 pKa = 3.38LYY261 pKa = 11.34SRR263 pKa = 11.84FVAQQPISEE272 pKa = 4.88PDD274 pKa = 3.66FLSQPSRR281 pKa = 11.84LVQFEE286 pKa = 4.09YY287 pKa = 10.7RR288 pKa = 11.84NPAFDD293 pKa = 5.21PDD295 pKa = 3.26VSLYY299 pKa = 10.11FEE301 pKa = 5.6RR302 pKa = 11.84DD303 pKa = 3.35LEE305 pKa = 4.3GLRR308 pKa = 11.84AAPLQEE314 pKa = 4.25FADD317 pKa = 3.91VVYY320 pKa = 10.67LGRR323 pKa = 11.84PRR325 pKa = 11.84VSSTSEE331 pKa = 3.36GTIRR335 pKa = 11.84VSRR338 pKa = 11.84LGTRR342 pKa = 11.84AALTTRR348 pKa = 11.84SGLSVGPQVHH358 pKa = 6.93FYY360 pKa = 10.1MDD362 pKa = 4.23LSDD365 pKa = 5.09IPPEE369 pKa = 4.17DD370 pKa = 4.13SIEE373 pKa = 4.11LHH375 pKa = 5.85TLNVTPQTSTIVDD388 pKa = 4.75DD389 pKa = 4.3ILATTTFDD397 pKa = 4.7DD398 pKa = 4.35PANSLFTQFNEE409 pKa = 4.93DD410 pKa = 3.56VLTDD414 pKa = 3.82DD415 pKa = 4.17VEE417 pKa = 5.22HH418 pKa = 6.78NFTEE422 pKa = 4.15SHH424 pKa = 6.0LVIPATDD431 pKa = 3.86EE432 pKa = 4.22EE433 pKa = 5.02NDD435 pKa = 3.6TAINIINLRR444 pKa = 11.84NIPLTVGMNSGDD456 pKa = 3.17ISTTLSDD463 pKa = 4.17YY464 pKa = 11.3NILDD468 pKa = 3.31ASLIVKK474 pKa = 10.29SNVSEE479 pKa = 4.08QPLFVLDD486 pKa = 3.7YY487 pKa = 11.05SDD489 pKa = 5.27YY490 pKa = 11.15DD491 pKa = 3.52LHH493 pKa = 7.29PGLLPKK499 pKa = 10.12RR500 pKa = 11.84RR501 pKa = 11.84RR502 pKa = 11.84IDD504 pKa = 3.38YY505 pKa = 9.4FF506 pKa = 3.72
MM1 pKa = 7.5LRR3 pKa = 11.84RR4 pKa = 11.84RR5 pKa = 11.84KK6 pKa = 9.54RR7 pKa = 11.84ASPTDD12 pKa = 4.02LYY14 pKa = 10.94RR15 pKa = 11.84SCLQGGDD22 pKa = 4.9CIPDD26 pKa = 3.37VQNKK30 pKa = 9.65FEE32 pKa = 4.96GNTIADD38 pKa = 3.83WLLKK42 pKa = 10.38IFGGLVYY49 pKa = 10.47FGNLGIGTGRR59 pKa = 11.84GTGGTFGYY67 pKa = 10.22RR68 pKa = 11.84PFGAPGSGRR77 pKa = 11.84PTQEE81 pKa = 4.06LPIARR86 pKa = 11.84PNVVIDD92 pKa = 4.1PLGPAPIVPVDD103 pKa = 3.96PSAASIVPLVEE114 pKa = 4.01GAPDD118 pKa = 3.39VGFAAPDD125 pKa = 3.7AGPAAGGTDD134 pKa = 2.95IEE136 pKa = 5.77LYY138 pKa = 10.36TITNSTTDD146 pKa = 3.08VGAVGGGPTVTSNEE160 pKa = 4.03EE161 pKa = 3.81FEE163 pKa = 4.65VAVIDD168 pKa = 4.12AQPIAPYY175 pKa = 9.75PKK177 pKa = 9.61QLLYY181 pKa = 11.04DD182 pKa = 3.6STIAATFEE190 pKa = 4.2TQINPFINPDD200 pKa = 3.17INNVNVLVDD209 pKa = 3.59PSFAGDD215 pKa = 3.44TVGDD219 pKa = 3.75YY220 pKa = 10.51FYY222 pKa = 11.5EE223 pKa = 4.62EE224 pKa = 4.36IPLEE228 pKa = 4.14RR229 pKa = 11.84LDD231 pKa = 3.68IQTFDD236 pKa = 3.1ILEE239 pKa = 4.72PPTEE243 pKa = 4.22STPTQLGNRR252 pKa = 11.84FVSRR256 pKa = 11.84ARR258 pKa = 11.84DD259 pKa = 3.38LYY261 pKa = 11.34SRR263 pKa = 11.84FVAQQPISEE272 pKa = 4.88PDD274 pKa = 3.66FLSQPSRR281 pKa = 11.84LVQFEE286 pKa = 4.09YY287 pKa = 10.7RR288 pKa = 11.84NPAFDD293 pKa = 5.21PDD295 pKa = 3.26VSLYY299 pKa = 10.11FEE301 pKa = 5.6RR302 pKa = 11.84DD303 pKa = 3.35LEE305 pKa = 4.3GLRR308 pKa = 11.84AAPLQEE314 pKa = 4.25FADD317 pKa = 3.91VVYY320 pKa = 10.67LGRR323 pKa = 11.84PRR325 pKa = 11.84VSSTSEE331 pKa = 3.36GTIRR335 pKa = 11.84VSRR338 pKa = 11.84LGTRR342 pKa = 11.84AALTTRR348 pKa = 11.84SGLSVGPQVHH358 pKa = 6.93FYY360 pKa = 10.1MDD362 pKa = 4.23LSDD365 pKa = 5.09IPPEE369 pKa = 4.17DD370 pKa = 4.13SIEE373 pKa = 4.11LHH375 pKa = 5.85TLNVTPQTSTIVDD388 pKa = 4.75DD389 pKa = 4.3ILATTTFDD397 pKa = 4.7DD398 pKa = 4.35PANSLFTQFNEE409 pKa = 4.93DD410 pKa = 3.56VLTDD414 pKa = 3.82DD415 pKa = 4.17VEE417 pKa = 5.22HH418 pKa = 6.78NFTEE422 pKa = 4.15SHH424 pKa = 6.0LVIPATDD431 pKa = 3.86EE432 pKa = 4.22EE433 pKa = 5.02NDD435 pKa = 3.6TAINIINLRR444 pKa = 11.84NIPLTVGMNSGDD456 pKa = 3.17ISTTLSDD463 pKa = 4.17YY464 pKa = 11.3NILDD468 pKa = 3.31ASLIVKK474 pKa = 10.29SNVSEE479 pKa = 4.08QPLFVLDD486 pKa = 3.7YY487 pKa = 11.05SDD489 pKa = 5.27YY490 pKa = 11.15DD491 pKa = 3.52LHH493 pKa = 7.29PGLLPKK499 pKa = 10.12RR500 pKa = 11.84RR501 pKa = 11.84RR502 pKa = 11.84IDD504 pKa = 3.38YY505 pKa = 9.4FF506 pKa = 3.72
Molecular weight: 55.2 kDa
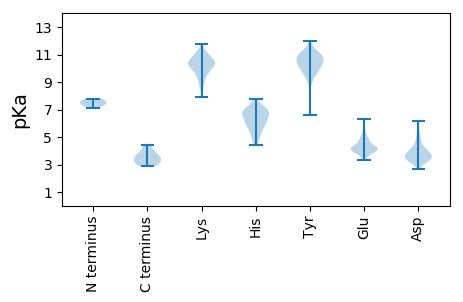
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q80931|Q80931_HPV50 E4 protein (Fragment) OS=Human papillomavirus type 50 OX=40539 GN=E4 PE=4 SV=1
MM1 pKa = 7.08TQMEE5 pKa = 4.44TQEE8 pKa = 4.16TLSARR13 pKa = 11.84FLAQQDD19 pKa = 3.2IQLNLIEE26 pKa = 5.45KK27 pKa = 10.09DD28 pKa = 3.64SKK30 pKa = 11.03NLKK33 pKa = 10.05DD34 pKa = 5.23HH35 pKa = 6.45IDD37 pKa = 3.31YY38 pKa = 9.76WEE40 pKa = 4.21SMRR43 pKa = 11.84KK44 pKa = 8.0EE45 pKa = 3.87QVLAFYY51 pKa = 10.55AKK53 pKa = 9.87KK54 pKa = 10.77EE55 pKa = 3.96NMSRR59 pKa = 11.84LGLQPLPPAKK69 pKa = 10.43VSEE72 pKa = 4.34QKK74 pKa = 11.03AKK76 pKa = 10.72DD77 pKa = 3.62AIRR80 pKa = 11.84IQLLLQSLYY89 pKa = 10.89KK90 pKa = 10.18SDD92 pKa = 4.33FGSEE96 pKa = 3.58PWTLSEE102 pKa = 5.6CSLEE106 pKa = 4.18MLNAPPRR113 pKa = 11.84NCFKK117 pKa = 10.61KK118 pKa = 10.32QPFTVTVQFDD128 pKa = 3.84NDD130 pKa = 3.74PKK132 pKa = 11.0NVYY135 pKa = 9.96PYY137 pKa = 9.92ICYY140 pKa = 10.19EE141 pKa = 3.92YY142 pKa = 10.03IYY144 pKa = 10.61YY145 pKa = 10.07QDD147 pKa = 6.16DD148 pKa = 3.18RR149 pKa = 11.84DD150 pKa = 3.42KK151 pKa = 8.74WHH153 pKa = 6.57KK154 pKa = 10.77VKK156 pKa = 10.88GLVDD160 pKa = 3.59HH161 pKa = 6.88NGLYY165 pKa = 10.41FKK167 pKa = 10.47EE168 pKa = 4.27VTGDD172 pKa = 3.27SVYY175 pKa = 10.94FKK177 pKa = 10.78LFQPDD182 pKa = 2.79ATVYY186 pKa = 10.3GKK188 pKa = 10.21SGQWTVIFKK197 pKa = 10.89NKK199 pKa = 8.9TIHH202 pKa = 6.79SSVTSSSRR210 pKa = 11.84SAFGPADD217 pKa = 3.93EE218 pKa = 4.78QPGPSTSYY226 pKa = 11.11DD227 pKa = 2.92KK228 pKa = 11.24SQQEE232 pKa = 4.35RR233 pKa = 11.84SGSGQPKK240 pKa = 9.85ALQDD244 pKa = 3.84TEE246 pKa = 4.83PPTSTSTVRR255 pKa = 11.84LRR257 pKa = 11.84RR258 pKa = 11.84GRR260 pKa = 11.84RR261 pKa = 11.84EE262 pKa = 4.1RR263 pKa = 11.84EE264 pKa = 3.67HH265 pKa = 6.93HH266 pKa = 6.34SYY268 pKa = 10.57RR269 pKa = 11.84HH270 pKa = 5.94RR271 pKa = 11.84KK272 pKa = 7.92SQSEE276 pKa = 4.24LGADD280 pKa = 3.54SAPTPEE286 pKa = 4.02EE287 pKa = 3.82VGRR290 pKa = 11.84RR291 pKa = 11.84SHH293 pKa = 5.88TVAAHH298 pKa = 5.29GLSRR302 pKa = 11.84LRR304 pKa = 11.84RR305 pKa = 11.84LQEE308 pKa = 3.72EE309 pKa = 4.14ARR311 pKa = 11.84DD312 pKa = 3.92PPVLIITGQQNNLKK326 pKa = 8.62CWRR329 pKa = 11.84YY330 pKa = 9.96RR331 pKa = 11.84FSQKK335 pKa = 10.54YY336 pKa = 9.1ADD338 pKa = 4.93LYY340 pKa = 9.51EE341 pKa = 4.74CCSSAWKK348 pKa = 9.36WLGPKK353 pKa = 9.87SEE355 pKa = 4.6GYY357 pKa = 10.35RR358 pKa = 11.84GDD360 pKa = 3.68AKK362 pKa = 11.16LLIAFKK368 pKa = 10.83NPEE371 pKa = 3.74QRR373 pKa = 11.84LSFLNTVGLPKK384 pKa = 9.9NTTYY388 pKa = 11.71SMGHH392 pKa = 6.95LDD394 pKa = 3.82SLL396 pKa = 4.39
MM1 pKa = 7.08TQMEE5 pKa = 4.44TQEE8 pKa = 4.16TLSARR13 pKa = 11.84FLAQQDD19 pKa = 3.2IQLNLIEE26 pKa = 5.45KK27 pKa = 10.09DD28 pKa = 3.64SKK30 pKa = 11.03NLKK33 pKa = 10.05DD34 pKa = 5.23HH35 pKa = 6.45IDD37 pKa = 3.31YY38 pKa = 9.76WEE40 pKa = 4.21SMRR43 pKa = 11.84KK44 pKa = 8.0EE45 pKa = 3.87QVLAFYY51 pKa = 10.55AKK53 pKa = 9.87KK54 pKa = 10.77EE55 pKa = 3.96NMSRR59 pKa = 11.84LGLQPLPPAKK69 pKa = 10.43VSEE72 pKa = 4.34QKK74 pKa = 11.03AKK76 pKa = 10.72DD77 pKa = 3.62AIRR80 pKa = 11.84IQLLLQSLYY89 pKa = 10.89KK90 pKa = 10.18SDD92 pKa = 4.33FGSEE96 pKa = 3.58PWTLSEE102 pKa = 5.6CSLEE106 pKa = 4.18MLNAPPRR113 pKa = 11.84NCFKK117 pKa = 10.61KK118 pKa = 10.32QPFTVTVQFDD128 pKa = 3.84NDD130 pKa = 3.74PKK132 pKa = 11.0NVYY135 pKa = 9.96PYY137 pKa = 9.92ICYY140 pKa = 10.19EE141 pKa = 3.92YY142 pKa = 10.03IYY144 pKa = 10.61YY145 pKa = 10.07QDD147 pKa = 6.16DD148 pKa = 3.18RR149 pKa = 11.84DD150 pKa = 3.42KK151 pKa = 8.74WHH153 pKa = 6.57KK154 pKa = 10.77VKK156 pKa = 10.88GLVDD160 pKa = 3.59HH161 pKa = 6.88NGLYY165 pKa = 10.41FKK167 pKa = 10.47EE168 pKa = 4.27VTGDD172 pKa = 3.27SVYY175 pKa = 10.94FKK177 pKa = 10.78LFQPDD182 pKa = 2.79ATVYY186 pKa = 10.3GKK188 pKa = 10.21SGQWTVIFKK197 pKa = 10.89NKK199 pKa = 8.9TIHH202 pKa = 6.79SSVTSSSRR210 pKa = 11.84SAFGPADD217 pKa = 3.93EE218 pKa = 4.78QPGPSTSYY226 pKa = 11.11DD227 pKa = 2.92KK228 pKa = 11.24SQQEE232 pKa = 4.35RR233 pKa = 11.84SGSGQPKK240 pKa = 9.85ALQDD244 pKa = 3.84TEE246 pKa = 4.83PPTSTSTVRR255 pKa = 11.84LRR257 pKa = 11.84RR258 pKa = 11.84GRR260 pKa = 11.84RR261 pKa = 11.84EE262 pKa = 4.1RR263 pKa = 11.84EE264 pKa = 3.67HH265 pKa = 6.93HH266 pKa = 6.34SYY268 pKa = 10.57RR269 pKa = 11.84HH270 pKa = 5.94RR271 pKa = 11.84KK272 pKa = 7.92SQSEE276 pKa = 4.24LGADD280 pKa = 3.54SAPTPEE286 pKa = 4.02EE287 pKa = 3.82VGRR290 pKa = 11.84RR291 pKa = 11.84SHH293 pKa = 5.88TVAAHH298 pKa = 5.29GLSRR302 pKa = 11.84LRR304 pKa = 11.84RR305 pKa = 11.84LQEE308 pKa = 3.72EE309 pKa = 4.14ARR311 pKa = 11.84DD312 pKa = 3.92PPVLIITGQQNNLKK326 pKa = 8.62CWRR329 pKa = 11.84YY330 pKa = 9.96RR331 pKa = 11.84FSQKK335 pKa = 10.54YY336 pKa = 9.1ADD338 pKa = 4.93LYY340 pKa = 9.51EE341 pKa = 4.74CCSSAWKK348 pKa = 9.36WLGPKK353 pKa = 9.87SEE355 pKa = 4.6GYY357 pKa = 10.35RR358 pKa = 11.84GDD360 pKa = 3.68AKK362 pKa = 11.16LLIAFKK368 pKa = 10.83NPEE371 pKa = 3.74QRR373 pKa = 11.84LSFLNTVGLPKK384 pKa = 9.9NTTYY388 pKa = 11.71SMGHH392 pKa = 6.95LDD394 pKa = 3.82SLL396 pKa = 4.39
Molecular weight: 45.56 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2396 |
93 |
599 |
342.3 |
38.87 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.676 ± 0.512 | 2.546 ± 0.814 |
7.137 ± 0.446 | 5.801 ± 0.356 |
4.633 ± 0.436 | 5.134 ± 0.89 |
1.92 ± 0.295 | 5.008 ± 0.606 |
5.593 ± 1.158 | 9.683 ± 0.687 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.586 ± 0.43 | 5.634 ± 0.642 |
5.843 ± 0.882 | 4.215 ± 0.596 |
5.217 ± 0.548 | 7.095 ± 0.52 |
6.01 ± 0.549 | 5.718 ± 0.675 |
1.169 ± 0.276 | 4.382 ± 0.483 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
