
Red clover cryptic virus 2
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Duplopiviricetes; Durnavirales; Partitiviridae; Betapartitivirus
Average proteome isoelectric point is 5.85
Get precalculated fractions of proteins
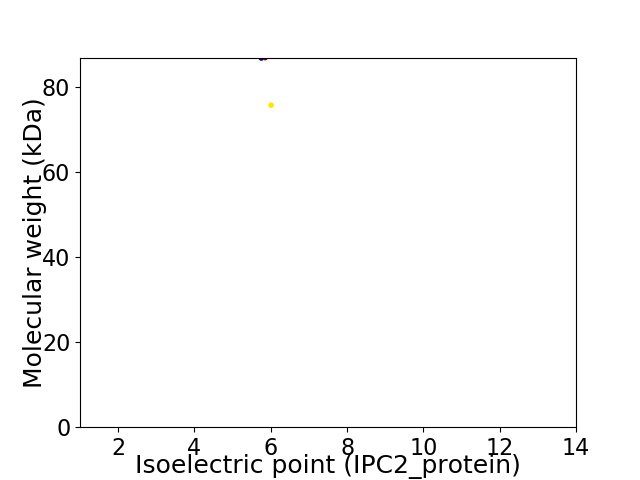
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|M9VYE7|M9VYE7_9VIRU Coat protein OS=Red clover cryptic virus 2 OX=1323524 PE=4 SV=1
MM1 pKa = 7.78PFNSARR7 pKa = 11.84NYY9 pKa = 9.61LAEE12 pKa = 4.08RR13 pKa = 11.84MIRR16 pKa = 11.84TKK18 pKa = 10.58QEE20 pKa = 3.28LMTYY24 pKa = 9.46QSEE27 pKa = 4.35DD28 pKa = 3.65HH29 pKa = 6.85NPDD32 pKa = 4.36AILEE36 pKa = 4.13KK37 pKa = 10.91SQDD40 pKa = 3.15PDD42 pKa = 3.03YY43 pKa = 11.22RR44 pKa = 11.84RR45 pKa = 11.84YY46 pKa = 10.46YY47 pKa = 11.06DD48 pKa = 3.2NTRR51 pKa = 11.84FDD53 pKa = 3.77PSNEE57 pKa = 3.79VKK59 pKa = 10.57HH60 pKa = 6.69RR61 pKa = 11.84ILNKK65 pKa = 9.87EE66 pKa = 3.61YY67 pKa = 8.91STLVEE72 pKa = 4.63AYY74 pKa = 10.04RR75 pKa = 11.84IKK77 pKa = 10.69NDD79 pKa = 3.74RR80 pKa = 11.84KK81 pKa = 7.94HH82 pKa = 5.81QPYY85 pKa = 9.82EE86 pKa = 3.6LHH88 pKa = 6.34QPIPEE93 pKa = 4.4DD94 pKa = 3.18AAPIPEE100 pKa = 4.3SRR102 pKa = 11.84VPAPGLKK109 pKa = 9.71LVPLMYY115 pKa = 10.13HH116 pKa = 6.24YY117 pKa = 11.17GHH119 pKa = 7.04IVHH122 pKa = 7.24DD123 pKa = 4.53PVASEE128 pKa = 5.03SDD130 pKa = 3.45SDD132 pKa = 4.29DD133 pKa = 5.05DD134 pKa = 3.76NTASEE139 pKa = 4.76RR140 pKa = 11.84PSKK143 pKa = 9.96TSIPHH148 pKa = 5.97FGYY151 pKa = 9.76PVNKK155 pKa = 9.71RR156 pKa = 11.84IYY158 pKa = 9.18DD159 pKa = 3.82VIVNVYY165 pKa = 9.8PEE167 pKa = 4.02YY168 pKa = 10.91LKK170 pKa = 11.18VIGEE174 pKa = 4.18YY175 pKa = 10.04CRR177 pKa = 11.84PIGTVEE183 pKa = 3.69ATFADD188 pKa = 4.64FNKK191 pKa = 9.91EE192 pKa = 4.12QIPSAPINIEE202 pKa = 3.67RR203 pKa = 11.84KK204 pKa = 8.71EE205 pKa = 3.92QVLTHH210 pKa = 6.44IFKK213 pKa = 10.77FLDD216 pKa = 3.75AQPYY220 pKa = 9.73LPLHH224 pKa = 5.98FVDD227 pKa = 4.11TQFCKK232 pKa = 10.39TPLVTGTGYY241 pKa = 10.81HH242 pKa = 5.59NRR244 pKa = 11.84YY245 pKa = 9.47SFKK248 pKa = 10.71QKK250 pKa = 10.44AHH252 pKa = 6.02AKK254 pKa = 8.37YY255 pKa = 10.37SRR257 pKa = 11.84PEE259 pKa = 3.92EE260 pKa = 4.18YY261 pKa = 10.83AKK263 pKa = 10.98LPTSKK268 pKa = 10.77GYY270 pKa = 10.32FYY272 pKa = 11.19NATYY276 pKa = 10.79EE277 pKa = 3.99NARR280 pKa = 11.84TLVHH284 pKa = 7.13FIKK287 pKa = 10.82QFGLPFNLQYY297 pKa = 11.12APEE300 pKa = 4.85DD301 pKa = 3.76ADD303 pKa = 3.67PTDD306 pKa = 3.97EE307 pKa = 4.44QVQSYY312 pKa = 10.59IDD314 pKa = 3.46TANSFFNDD322 pKa = 3.36YY323 pKa = 7.74PTLLFTRR330 pKa = 11.84NHH332 pKa = 5.43ISKK335 pKa = 10.37RR336 pKa = 11.84DD337 pKa = 3.44GTLKK341 pKa = 10.16VRR343 pKa = 11.84PVYY346 pKa = 10.61AVDD349 pKa = 3.86DD350 pKa = 4.04LFIIIEE356 pKa = 4.15LMLTFPLTVQARR368 pKa = 11.84KK369 pKa = 9.5QSCCIMYY376 pKa = 10.47GLEE379 pKa = 4.17TIRR382 pKa = 11.84GSNHH386 pKa = 5.57YY387 pKa = 9.72IEE389 pKa = 5.15RR390 pKa = 11.84LARR393 pKa = 11.84SYY395 pKa = 10.05STYY398 pKa = 10.64FSLDD402 pKa = 2.68WSSYY406 pKa = 6.99DD407 pKa = 3.17QRR409 pKa = 11.84LPRR412 pKa = 11.84VITDD416 pKa = 3.3IYY418 pKa = 9.53YY419 pKa = 9.37TDD421 pKa = 3.94FLRR424 pKa = 11.84RR425 pKa = 11.84LIVINHH431 pKa = 7.13GYY433 pKa = 9.18QPTYY437 pKa = 10.39EE438 pKa = 4.22YY439 pKa = 8.37PTYY442 pKa = 10.44PDD444 pKa = 3.72LDD446 pKa = 3.56EE447 pKa = 5.2HH448 pKa = 7.08KK449 pKa = 10.58LYY451 pKa = 11.33SRR453 pKa = 11.84MNNLLYY459 pKa = 10.34FLHH462 pKa = 6.11TWYY465 pKa = 11.33NNMTFVLSDD474 pKa = 3.4GYY476 pKa = 11.22AYY478 pKa = 10.65RR479 pKa = 11.84RR480 pKa = 11.84THH482 pKa = 6.4CGVPSGLYY490 pKa = 7.56NTQYY494 pKa = 11.31LDD496 pKa = 3.35SFGNLFLIIDD506 pKa = 4.03AMLEE510 pKa = 4.09FGFSEE515 pKa = 4.57SEE517 pKa = 3.34IDD519 pKa = 4.26NFILLVLGDD528 pKa = 5.24DD529 pKa = 3.95NTGMTVISIDD539 pKa = 4.12RR540 pKa = 11.84IYY542 pKa = 11.38DD543 pKa = 3.97FINFLEE549 pKa = 4.62KK550 pKa = 10.68YY551 pKa = 10.59ALIRR555 pKa = 11.84YY556 pKa = 8.82NMVLSPTKK564 pKa = 10.56SVLTTLRR571 pKa = 11.84SKK573 pKa = 10.73IEE575 pKa = 3.91TLGYY579 pKa = 9.27QCNHH583 pKa = 6.68GSPKK587 pKa = 9.95RR588 pKa = 11.84DD589 pKa = 2.91ISKK592 pKa = 10.59LVAQLCYY599 pKa = 9.98PEE601 pKa = 5.79NGLKK605 pKa = 9.45PHH607 pKa = 6.12TMAARR612 pKa = 11.84AIGIAYY618 pKa = 8.9AAAGQDD624 pKa = 3.48PMFHH628 pKa = 6.27SFCHH632 pKa = 6.17DD633 pKa = 3.26VYY635 pKa = 11.72NLFRR639 pKa = 11.84LDD641 pKa = 3.9YY642 pKa = 10.95KK643 pKa = 10.9PDD645 pKa = 3.05ARR647 pKa = 11.84TNLNFQRR654 pKa = 11.84QIYY657 pKa = 9.6HH658 pKa = 6.09NLEE661 pKa = 4.32DD662 pKa = 5.48GIPDD666 pKa = 4.25LATPVVPPFPSLYY679 pKa = 9.46EE680 pKa = 3.88VRR682 pKa = 11.84HH683 pKa = 5.87MYY685 pKa = 10.45SKK687 pKa = 10.62YY688 pKa = 10.07QGPLSYY694 pKa = 10.73APKK697 pKa = 9.2WNYY700 pKa = 10.18AHH702 pKa = 7.53FINDD706 pKa = 3.81PDD708 pKa = 3.9VTPPSPKK715 pKa = 9.65TMRR718 pKa = 11.84DD719 pKa = 3.43YY720 pKa = 11.07EE721 pKa = 4.24IEE723 pKa = 3.97NDD725 pKa = 4.35LISRR729 pKa = 11.84TAPTFEE735 pKa = 4.15TVVPATRR742 pKa = 11.84NFPP745 pKa = 3.73
MM1 pKa = 7.78PFNSARR7 pKa = 11.84NYY9 pKa = 9.61LAEE12 pKa = 4.08RR13 pKa = 11.84MIRR16 pKa = 11.84TKK18 pKa = 10.58QEE20 pKa = 3.28LMTYY24 pKa = 9.46QSEE27 pKa = 4.35DD28 pKa = 3.65HH29 pKa = 6.85NPDD32 pKa = 4.36AILEE36 pKa = 4.13KK37 pKa = 10.91SQDD40 pKa = 3.15PDD42 pKa = 3.03YY43 pKa = 11.22RR44 pKa = 11.84RR45 pKa = 11.84YY46 pKa = 10.46YY47 pKa = 11.06DD48 pKa = 3.2NTRR51 pKa = 11.84FDD53 pKa = 3.77PSNEE57 pKa = 3.79VKK59 pKa = 10.57HH60 pKa = 6.69RR61 pKa = 11.84ILNKK65 pKa = 9.87EE66 pKa = 3.61YY67 pKa = 8.91STLVEE72 pKa = 4.63AYY74 pKa = 10.04RR75 pKa = 11.84IKK77 pKa = 10.69NDD79 pKa = 3.74RR80 pKa = 11.84KK81 pKa = 7.94HH82 pKa = 5.81QPYY85 pKa = 9.82EE86 pKa = 3.6LHH88 pKa = 6.34QPIPEE93 pKa = 4.4DD94 pKa = 3.18AAPIPEE100 pKa = 4.3SRR102 pKa = 11.84VPAPGLKK109 pKa = 9.71LVPLMYY115 pKa = 10.13HH116 pKa = 6.24YY117 pKa = 11.17GHH119 pKa = 7.04IVHH122 pKa = 7.24DD123 pKa = 4.53PVASEE128 pKa = 5.03SDD130 pKa = 3.45SDD132 pKa = 4.29DD133 pKa = 5.05DD134 pKa = 3.76NTASEE139 pKa = 4.76RR140 pKa = 11.84PSKK143 pKa = 9.96TSIPHH148 pKa = 5.97FGYY151 pKa = 9.76PVNKK155 pKa = 9.71RR156 pKa = 11.84IYY158 pKa = 9.18DD159 pKa = 3.82VIVNVYY165 pKa = 9.8PEE167 pKa = 4.02YY168 pKa = 10.91LKK170 pKa = 11.18VIGEE174 pKa = 4.18YY175 pKa = 10.04CRR177 pKa = 11.84PIGTVEE183 pKa = 3.69ATFADD188 pKa = 4.64FNKK191 pKa = 9.91EE192 pKa = 4.12QIPSAPINIEE202 pKa = 3.67RR203 pKa = 11.84KK204 pKa = 8.71EE205 pKa = 3.92QVLTHH210 pKa = 6.44IFKK213 pKa = 10.77FLDD216 pKa = 3.75AQPYY220 pKa = 9.73LPLHH224 pKa = 5.98FVDD227 pKa = 4.11TQFCKK232 pKa = 10.39TPLVTGTGYY241 pKa = 10.81HH242 pKa = 5.59NRR244 pKa = 11.84YY245 pKa = 9.47SFKK248 pKa = 10.71QKK250 pKa = 10.44AHH252 pKa = 6.02AKK254 pKa = 8.37YY255 pKa = 10.37SRR257 pKa = 11.84PEE259 pKa = 3.92EE260 pKa = 4.18YY261 pKa = 10.83AKK263 pKa = 10.98LPTSKK268 pKa = 10.77GYY270 pKa = 10.32FYY272 pKa = 11.19NATYY276 pKa = 10.79EE277 pKa = 3.99NARR280 pKa = 11.84TLVHH284 pKa = 7.13FIKK287 pKa = 10.82QFGLPFNLQYY297 pKa = 11.12APEE300 pKa = 4.85DD301 pKa = 3.76ADD303 pKa = 3.67PTDD306 pKa = 3.97EE307 pKa = 4.44QVQSYY312 pKa = 10.59IDD314 pKa = 3.46TANSFFNDD322 pKa = 3.36YY323 pKa = 7.74PTLLFTRR330 pKa = 11.84NHH332 pKa = 5.43ISKK335 pKa = 10.37RR336 pKa = 11.84DD337 pKa = 3.44GTLKK341 pKa = 10.16VRR343 pKa = 11.84PVYY346 pKa = 10.61AVDD349 pKa = 3.86DD350 pKa = 4.04LFIIIEE356 pKa = 4.15LMLTFPLTVQARR368 pKa = 11.84KK369 pKa = 9.5QSCCIMYY376 pKa = 10.47GLEE379 pKa = 4.17TIRR382 pKa = 11.84GSNHH386 pKa = 5.57YY387 pKa = 9.72IEE389 pKa = 5.15RR390 pKa = 11.84LARR393 pKa = 11.84SYY395 pKa = 10.05STYY398 pKa = 10.64FSLDD402 pKa = 2.68WSSYY406 pKa = 6.99DD407 pKa = 3.17QRR409 pKa = 11.84LPRR412 pKa = 11.84VITDD416 pKa = 3.3IYY418 pKa = 9.53YY419 pKa = 9.37TDD421 pKa = 3.94FLRR424 pKa = 11.84RR425 pKa = 11.84LIVINHH431 pKa = 7.13GYY433 pKa = 9.18QPTYY437 pKa = 10.39EE438 pKa = 4.22YY439 pKa = 8.37PTYY442 pKa = 10.44PDD444 pKa = 3.72LDD446 pKa = 3.56EE447 pKa = 5.2HH448 pKa = 7.08KK449 pKa = 10.58LYY451 pKa = 11.33SRR453 pKa = 11.84MNNLLYY459 pKa = 10.34FLHH462 pKa = 6.11TWYY465 pKa = 11.33NNMTFVLSDD474 pKa = 3.4GYY476 pKa = 11.22AYY478 pKa = 10.65RR479 pKa = 11.84RR480 pKa = 11.84THH482 pKa = 6.4CGVPSGLYY490 pKa = 7.56NTQYY494 pKa = 11.31LDD496 pKa = 3.35SFGNLFLIIDD506 pKa = 4.03AMLEE510 pKa = 4.09FGFSEE515 pKa = 4.57SEE517 pKa = 3.34IDD519 pKa = 4.26NFILLVLGDD528 pKa = 5.24DD529 pKa = 3.95NTGMTVISIDD539 pKa = 4.12RR540 pKa = 11.84IYY542 pKa = 11.38DD543 pKa = 3.97FINFLEE549 pKa = 4.62KK550 pKa = 10.68YY551 pKa = 10.59ALIRR555 pKa = 11.84YY556 pKa = 8.82NMVLSPTKK564 pKa = 10.56SVLTTLRR571 pKa = 11.84SKK573 pKa = 10.73IEE575 pKa = 3.91TLGYY579 pKa = 9.27QCNHH583 pKa = 6.68GSPKK587 pKa = 9.95RR588 pKa = 11.84DD589 pKa = 2.91ISKK592 pKa = 10.59LVAQLCYY599 pKa = 9.98PEE601 pKa = 5.79NGLKK605 pKa = 9.45PHH607 pKa = 6.12TMAARR612 pKa = 11.84AIGIAYY618 pKa = 8.9AAAGQDD624 pKa = 3.48PMFHH628 pKa = 6.27SFCHH632 pKa = 6.17DD633 pKa = 3.26VYY635 pKa = 11.72NLFRR639 pKa = 11.84LDD641 pKa = 3.9YY642 pKa = 10.95KK643 pKa = 10.9PDD645 pKa = 3.05ARR647 pKa = 11.84TNLNFQRR654 pKa = 11.84QIYY657 pKa = 9.6HH658 pKa = 6.09NLEE661 pKa = 4.32DD662 pKa = 5.48GIPDD666 pKa = 4.25LATPVVPPFPSLYY679 pKa = 9.46EE680 pKa = 3.88VRR682 pKa = 11.84HH683 pKa = 5.87MYY685 pKa = 10.45SKK687 pKa = 10.62YY688 pKa = 10.07QGPLSYY694 pKa = 10.73APKK697 pKa = 9.2WNYY700 pKa = 10.18AHH702 pKa = 7.53FINDD706 pKa = 3.81PDD708 pKa = 3.9VTPPSPKK715 pKa = 9.65TMRR718 pKa = 11.84DD719 pKa = 3.43YY720 pKa = 11.07EE721 pKa = 4.24IEE723 pKa = 3.97NDD725 pKa = 4.35LISRR729 pKa = 11.84TAPTFEE735 pKa = 4.15TVVPATRR742 pKa = 11.84NFPP745 pKa = 3.73
Molecular weight: 86.76 kDa
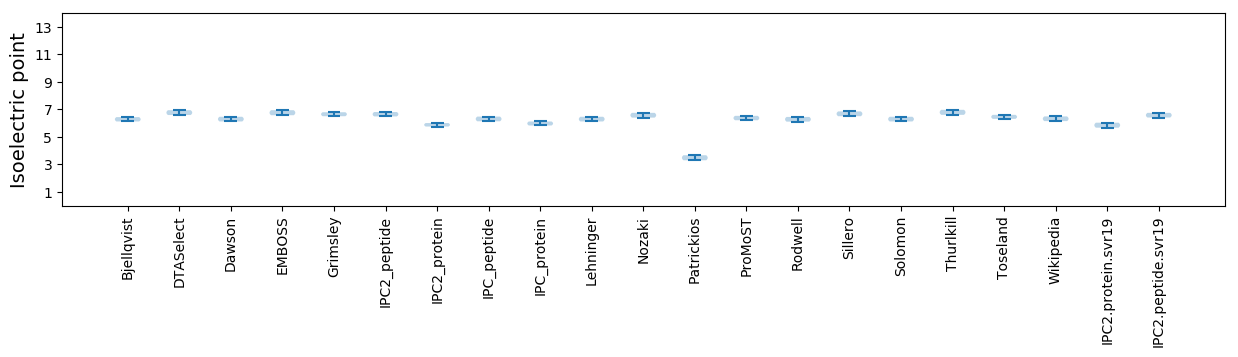
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|M9VYE7|M9VYE7_9VIRU Coat protein OS=Red clover cryptic virus 2 OX=1323524 PE=4 SV=1
MM1 pKa = 6.97STEE4 pKa = 3.85EE5 pKa = 4.24TLPPTKK11 pKa = 9.94TIKK14 pKa = 9.92NQNVNLIFQDD24 pKa = 3.51TKK26 pKa = 11.44VPIPSDD32 pKa = 2.97NFMEE36 pKa = 4.57LLRR39 pKa = 11.84SNAAAHH45 pKa = 6.57TDD47 pKa = 3.46TSNLWVIDD55 pKa = 3.56VLPNFTPILMFIMHH69 pKa = 7.06YY70 pKa = 10.9AHH72 pKa = 7.0IHH74 pKa = 5.2AQAADD79 pKa = 3.66YY80 pKa = 10.16RR81 pKa = 11.84VHH83 pKa = 6.82AKK85 pKa = 10.26SSAYY89 pKa = 7.55TLCMYY94 pKa = 11.06YY95 pKa = 7.67MTVVYY100 pKa = 9.74GYY102 pKa = 10.96FLLNDD107 pKa = 3.84LHH109 pKa = 6.1TRR111 pKa = 11.84PTRR114 pKa = 11.84SVHH117 pKa = 4.6ARR119 pKa = 11.84VWVEE123 pKa = 3.69SSWRR127 pKa = 11.84SDD129 pKa = 3.16FVKK132 pKa = 10.44FLTTLPVPEE141 pKa = 4.3FLKK144 pKa = 10.88PILSQFHH151 pKa = 6.96PFNTDD156 pKa = 2.27RR157 pKa = 11.84TKK159 pKa = 11.26NVFFSPSAAGFDD171 pKa = 3.32HH172 pKa = 7.28DD173 pKa = 4.43QFFGRR178 pKa = 11.84IFPMNMFAAFHH189 pKa = 7.31DD190 pKa = 4.61CTATLPGNSSKK201 pKa = 10.95VDD203 pKa = 3.41VLRR206 pKa = 11.84NLYY209 pKa = 8.6TRR211 pKa = 11.84NLYY214 pKa = 9.93TITAPGYY221 pKa = 7.97TCMIPDD227 pKa = 5.67LIGITPDD234 pKa = 2.81QAAATTVNYY243 pKa = 8.75MNSKK247 pKa = 10.02FYY249 pKa = 10.03QTFTSIFNPVLFRR262 pKa = 11.84DD263 pKa = 3.86SQRR266 pKa = 11.84RR267 pKa = 11.84ASLAALSFTAPTYY280 pKa = 7.37PTNHH284 pKa = 5.98INAYY288 pKa = 10.43DD289 pKa = 3.76YY290 pKa = 10.03MFSATAPNLRR300 pKa = 11.84EE301 pKa = 3.95LRR303 pKa = 11.84IVLQSIIAITKK314 pKa = 10.12DD315 pKa = 3.35VFKK318 pKa = 11.06SDD320 pKa = 3.61QTLGNYY326 pKa = 8.2VADD329 pKa = 3.93YY330 pKa = 10.55SSGSIIHH337 pKa = 6.59HH338 pKa = 6.77GYY340 pKa = 7.78STYY343 pKa = 11.42ALPTWSHH350 pKa = 6.09TPTTGKK356 pKa = 10.06FDD358 pKa = 3.91RR359 pKa = 11.84FNTANTFNLVTEE371 pKa = 4.51DD372 pKa = 4.02ARR374 pKa = 11.84AEE376 pKa = 4.5DD377 pKa = 3.42ICFLQRR383 pKa = 11.84PAAEE387 pKa = 4.24IPHH390 pKa = 6.24TRR392 pKa = 11.84LITDD396 pKa = 3.76IVYY399 pKa = 10.39AATTAPATSLTVPANHH415 pKa = 6.79ALYY418 pKa = 10.44RR419 pKa = 11.84RR420 pKa = 11.84WPYY423 pKa = 10.69CLRR426 pKa = 11.84LDD428 pKa = 3.75EE429 pKa = 5.17DD430 pKa = 4.07AANGFPRR437 pKa = 11.84HH438 pKa = 6.25DD439 pKa = 4.23NEE441 pKa = 4.89DD442 pKa = 3.34LVAFSAEE449 pKa = 3.73SHH451 pKa = 5.94AAPRR455 pKa = 11.84VLVLDD460 pKa = 4.05TPGDD464 pKa = 4.07TVTSAHH470 pKa = 6.69LPTLTGKK477 pKa = 10.53VIEE480 pKa = 4.35SFEE483 pKa = 4.76LDD485 pKa = 3.07GSTIEE490 pKa = 4.42MPDD493 pKa = 3.13ARR495 pKa = 11.84KK496 pKa = 10.04SLGMQNCMFADD507 pKa = 3.4SAIAYY512 pKa = 8.89KK513 pKa = 9.56YY514 pKa = 9.9VRR516 pKa = 11.84PGSYY520 pKa = 8.67WRR522 pKa = 11.84PRR524 pKa = 11.84AAGGTLPPLNRR535 pKa = 11.84APPNSRR541 pKa = 11.84PRR543 pKa = 11.84LPASSLLHH551 pKa = 6.91DD552 pKa = 3.74RR553 pKa = 11.84TKK555 pKa = 11.14VFLPQLNRR563 pKa = 11.84HH564 pKa = 5.06VNAVPDD570 pKa = 3.78NDD572 pKa = 4.04ALPGFTQITPVNVIRR587 pKa = 11.84YY588 pKa = 7.25IQSFLGFRR596 pKa = 11.84TVDD599 pKa = 3.14SSANEE604 pKa = 3.86AALDD608 pKa = 3.78AVPGMNEE615 pKa = 3.99SLLMIWSPYY624 pKa = 8.28TYY626 pKa = 10.77NPYY629 pKa = 10.77EE630 pKa = 4.54SDD632 pKa = 4.28DD633 pKa = 4.15YY634 pKa = 11.46PALAYY639 pKa = 9.46DD640 pKa = 4.28ASRR643 pKa = 11.84HH644 pKa = 5.13YY645 pKa = 11.23YY646 pKa = 8.29LTNLRR651 pKa = 11.84TIFGTDD657 pKa = 3.27YY658 pKa = 11.17NLVATKK664 pKa = 10.12HH665 pKa = 6.0IYY667 pKa = 10.35EE668 pKa = 4.55SFPAVV673 pKa = 3.04
MM1 pKa = 6.97STEE4 pKa = 3.85EE5 pKa = 4.24TLPPTKK11 pKa = 9.94TIKK14 pKa = 9.92NQNVNLIFQDD24 pKa = 3.51TKK26 pKa = 11.44VPIPSDD32 pKa = 2.97NFMEE36 pKa = 4.57LLRR39 pKa = 11.84SNAAAHH45 pKa = 6.57TDD47 pKa = 3.46TSNLWVIDD55 pKa = 3.56VLPNFTPILMFIMHH69 pKa = 7.06YY70 pKa = 10.9AHH72 pKa = 7.0IHH74 pKa = 5.2AQAADD79 pKa = 3.66YY80 pKa = 10.16RR81 pKa = 11.84VHH83 pKa = 6.82AKK85 pKa = 10.26SSAYY89 pKa = 7.55TLCMYY94 pKa = 11.06YY95 pKa = 7.67MTVVYY100 pKa = 9.74GYY102 pKa = 10.96FLLNDD107 pKa = 3.84LHH109 pKa = 6.1TRR111 pKa = 11.84PTRR114 pKa = 11.84SVHH117 pKa = 4.6ARR119 pKa = 11.84VWVEE123 pKa = 3.69SSWRR127 pKa = 11.84SDD129 pKa = 3.16FVKK132 pKa = 10.44FLTTLPVPEE141 pKa = 4.3FLKK144 pKa = 10.88PILSQFHH151 pKa = 6.96PFNTDD156 pKa = 2.27RR157 pKa = 11.84TKK159 pKa = 11.26NVFFSPSAAGFDD171 pKa = 3.32HH172 pKa = 7.28DD173 pKa = 4.43QFFGRR178 pKa = 11.84IFPMNMFAAFHH189 pKa = 7.31DD190 pKa = 4.61CTATLPGNSSKK201 pKa = 10.95VDD203 pKa = 3.41VLRR206 pKa = 11.84NLYY209 pKa = 8.6TRR211 pKa = 11.84NLYY214 pKa = 9.93TITAPGYY221 pKa = 7.97TCMIPDD227 pKa = 5.67LIGITPDD234 pKa = 2.81QAAATTVNYY243 pKa = 8.75MNSKK247 pKa = 10.02FYY249 pKa = 10.03QTFTSIFNPVLFRR262 pKa = 11.84DD263 pKa = 3.86SQRR266 pKa = 11.84RR267 pKa = 11.84ASLAALSFTAPTYY280 pKa = 7.37PTNHH284 pKa = 5.98INAYY288 pKa = 10.43DD289 pKa = 3.76YY290 pKa = 10.03MFSATAPNLRR300 pKa = 11.84EE301 pKa = 3.95LRR303 pKa = 11.84IVLQSIIAITKK314 pKa = 10.12DD315 pKa = 3.35VFKK318 pKa = 11.06SDD320 pKa = 3.61QTLGNYY326 pKa = 8.2VADD329 pKa = 3.93YY330 pKa = 10.55SSGSIIHH337 pKa = 6.59HH338 pKa = 6.77GYY340 pKa = 7.78STYY343 pKa = 11.42ALPTWSHH350 pKa = 6.09TPTTGKK356 pKa = 10.06FDD358 pKa = 3.91RR359 pKa = 11.84FNTANTFNLVTEE371 pKa = 4.51DD372 pKa = 4.02ARR374 pKa = 11.84AEE376 pKa = 4.5DD377 pKa = 3.42ICFLQRR383 pKa = 11.84PAAEE387 pKa = 4.24IPHH390 pKa = 6.24TRR392 pKa = 11.84LITDD396 pKa = 3.76IVYY399 pKa = 10.39AATTAPATSLTVPANHH415 pKa = 6.79ALYY418 pKa = 10.44RR419 pKa = 11.84RR420 pKa = 11.84WPYY423 pKa = 10.69CLRR426 pKa = 11.84LDD428 pKa = 3.75EE429 pKa = 5.17DD430 pKa = 4.07AANGFPRR437 pKa = 11.84HH438 pKa = 6.25DD439 pKa = 4.23NEE441 pKa = 4.89DD442 pKa = 3.34LVAFSAEE449 pKa = 3.73SHH451 pKa = 5.94AAPRR455 pKa = 11.84VLVLDD460 pKa = 4.05TPGDD464 pKa = 4.07TVTSAHH470 pKa = 6.69LPTLTGKK477 pKa = 10.53VIEE480 pKa = 4.35SFEE483 pKa = 4.76LDD485 pKa = 3.07GSTIEE490 pKa = 4.42MPDD493 pKa = 3.13ARR495 pKa = 11.84KK496 pKa = 10.04SLGMQNCMFADD507 pKa = 3.4SAIAYY512 pKa = 8.89KK513 pKa = 9.56YY514 pKa = 9.9VRR516 pKa = 11.84PGSYY520 pKa = 8.67WRR522 pKa = 11.84PRR524 pKa = 11.84AAGGTLPPLNRR535 pKa = 11.84APPNSRR541 pKa = 11.84PRR543 pKa = 11.84LPASSLLHH551 pKa = 6.91DD552 pKa = 3.74RR553 pKa = 11.84TKK555 pKa = 11.14VFLPQLNRR563 pKa = 11.84HH564 pKa = 5.06VNAVPDD570 pKa = 3.78NDD572 pKa = 4.04ALPGFTQITPVNVIRR587 pKa = 11.84YY588 pKa = 7.25IQSFLGFRR596 pKa = 11.84TVDD599 pKa = 3.14SSANEE604 pKa = 3.86AALDD608 pKa = 3.78AVPGMNEE615 pKa = 3.99SLLMIWSPYY624 pKa = 8.28TYY626 pKa = 10.77NPYY629 pKa = 10.77EE630 pKa = 4.54SDD632 pKa = 4.28DD633 pKa = 4.15YY634 pKa = 11.46PALAYY639 pKa = 9.46DD640 pKa = 4.28ASRR643 pKa = 11.84HH644 pKa = 5.13YY645 pKa = 11.23YY646 pKa = 8.29LTNLRR651 pKa = 11.84TIFGTDD657 pKa = 3.27YY658 pKa = 11.17NLVATKK664 pKa = 10.12HH665 pKa = 6.0IYY667 pKa = 10.35EE668 pKa = 4.55SFPAVV673 pKa = 3.04
Molecular weight: 75.68 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1418 |
673 |
745 |
709.0 |
81.22 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.546 ± 1.567 | 0.987 ± 0.066 |
6.417 ± 0.225 | 4.09 ± 0.878 |
5.43 ± 0.253 | 3.456 ± 0.129 |
3.456 ± 0.026 | 5.712 ± 0.457 |
3.597 ± 0.742 | 8.533 ± 0.059 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.186 ± 0.133 | 5.642 ± 0.106 |
7.475 ± 0.032 | 2.821 ± 0.41 |
5.642 ± 0.1 | 6.488 ± 0.446 |
7.828 ± 1.062 | 5.289 ± 0.248 |
0.705 ± 0.232 | 6.7 ± 1.039 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
