
Wenling sobemo-like virus 1
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 5.81
Get precalculated fractions of proteins
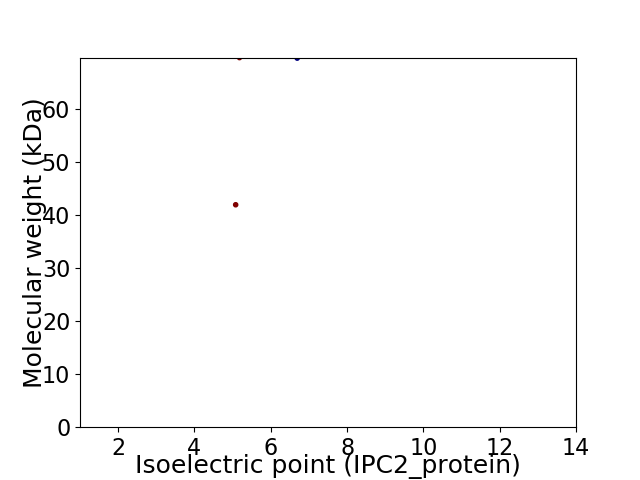
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KFA0|A0A1L3KFA0_9VIRU RNA-directed RNA polymerase OS=Wenling sobemo-like virus 1 OX=1923540 PE=4 SV=1
MM1 pKa = 6.87STLVSIVKK9 pKa = 10.38DD10 pKa = 3.51LVDD13 pKa = 3.43VEE15 pKa = 4.91NPVDD19 pKa = 4.33YY20 pKa = 7.62PTLDD24 pKa = 3.3WFNHH28 pKa = 4.71VLLHH32 pKa = 6.88DD33 pKa = 4.07VTPSSSTGLPWKK45 pKa = 10.1KK46 pKa = 10.19RR47 pKa = 11.84GMDD50 pKa = 3.18KK51 pKa = 10.97GSSFVEE57 pKa = 4.26FGEE60 pKa = 4.04IKK62 pKa = 10.45EE63 pKa = 4.31NAIHH67 pKa = 7.18LYY69 pKa = 9.95DD70 pKa = 3.7AVVSRR75 pKa = 11.84VLALSEE81 pKa = 4.25GVEE84 pKa = 4.4SDD86 pKa = 5.01PISCFIKK93 pKa = 10.54PEE95 pKa = 3.72PHH97 pKa = 6.66KK98 pKa = 10.29PSKK101 pKa = 10.94VKK103 pKa = 10.76DD104 pKa = 3.6GAWRR108 pKa = 11.84IISGVGITDD117 pKa = 3.85CFVDD121 pKa = 5.05RR122 pKa = 11.84MLFGNFLNGSIEE134 pKa = 4.09KK135 pKa = 10.59ARR137 pKa = 11.84TLDD140 pKa = 3.6NPLLPGFVPYY150 pKa = 10.65FGGYY154 pKa = 9.0RR155 pKa = 11.84DD156 pKa = 3.65LARR159 pKa = 11.84KK160 pKa = 8.68FVRR163 pKa = 11.84PEE165 pKa = 3.79TADD168 pKa = 3.25KK169 pKa = 11.24SSWDD173 pKa = 3.27WTVQGWMVTFLRR185 pKa = 11.84RR186 pKa = 11.84FFEE189 pKa = 4.46AVTDD193 pKa = 4.31SKK195 pKa = 10.85WHH197 pKa = 6.43GILNNRR203 pKa = 11.84LTALFQDD210 pKa = 3.96AEE212 pKa = 4.49LDD214 pKa = 3.69FEE216 pKa = 5.1GYY218 pKa = 10.24RR219 pKa = 11.84FRR221 pKa = 11.84QNCDD225 pKa = 3.09GIMKK229 pKa = 9.86SGCLGTIIFNSLLQLAIHH247 pKa = 6.18VLSCMRR253 pKa = 11.84SGEE256 pKa = 4.08DD257 pKa = 3.45LKK259 pKa = 11.14PFYY262 pKa = 10.9AVGDD266 pKa = 3.76DD267 pKa = 4.73TIQEE271 pKa = 4.42TCSEE275 pKa = 4.46EE276 pKa = 3.9YY277 pKa = 9.16WDD279 pKa = 3.94EE280 pKa = 4.15VKK282 pKa = 10.78KK283 pKa = 11.03CGAIVKK289 pKa = 9.2EE290 pKa = 4.9RR291 pKa = 11.84IGGWPAEE298 pKa = 4.15FIGIYY303 pKa = 10.68FNDD306 pKa = 4.0TVGLPMYY313 pKa = 8.17ATKK316 pKa = 10.98NFFRR320 pKa = 11.84LLFAEE325 pKa = 5.4AEE327 pKa = 4.14LLDD330 pKa = 5.71DD331 pKa = 4.27ILAGYY336 pKa = 7.89MRR338 pKa = 11.84LYY340 pKa = 11.05ANSPLFTILNEE351 pKa = 3.62LAIRR355 pKa = 11.84KK356 pKa = 8.39GVTLSKK362 pKa = 10.64DD363 pKa = 3.42DD364 pKa = 3.88CLTWFNGG371 pKa = 3.39
MM1 pKa = 6.87STLVSIVKK9 pKa = 10.38DD10 pKa = 3.51LVDD13 pKa = 3.43VEE15 pKa = 4.91NPVDD19 pKa = 4.33YY20 pKa = 7.62PTLDD24 pKa = 3.3WFNHH28 pKa = 4.71VLLHH32 pKa = 6.88DD33 pKa = 4.07VTPSSSTGLPWKK45 pKa = 10.1KK46 pKa = 10.19RR47 pKa = 11.84GMDD50 pKa = 3.18KK51 pKa = 10.97GSSFVEE57 pKa = 4.26FGEE60 pKa = 4.04IKK62 pKa = 10.45EE63 pKa = 4.31NAIHH67 pKa = 7.18LYY69 pKa = 9.95DD70 pKa = 3.7AVVSRR75 pKa = 11.84VLALSEE81 pKa = 4.25GVEE84 pKa = 4.4SDD86 pKa = 5.01PISCFIKK93 pKa = 10.54PEE95 pKa = 3.72PHH97 pKa = 6.66KK98 pKa = 10.29PSKK101 pKa = 10.94VKK103 pKa = 10.76DD104 pKa = 3.6GAWRR108 pKa = 11.84IISGVGITDD117 pKa = 3.85CFVDD121 pKa = 5.05RR122 pKa = 11.84MLFGNFLNGSIEE134 pKa = 4.09KK135 pKa = 10.59ARR137 pKa = 11.84TLDD140 pKa = 3.6NPLLPGFVPYY150 pKa = 10.65FGGYY154 pKa = 9.0RR155 pKa = 11.84DD156 pKa = 3.65LARR159 pKa = 11.84KK160 pKa = 8.68FVRR163 pKa = 11.84PEE165 pKa = 3.79TADD168 pKa = 3.25KK169 pKa = 11.24SSWDD173 pKa = 3.27WTVQGWMVTFLRR185 pKa = 11.84RR186 pKa = 11.84FFEE189 pKa = 4.46AVTDD193 pKa = 4.31SKK195 pKa = 10.85WHH197 pKa = 6.43GILNNRR203 pKa = 11.84LTALFQDD210 pKa = 3.96AEE212 pKa = 4.49LDD214 pKa = 3.69FEE216 pKa = 5.1GYY218 pKa = 10.24RR219 pKa = 11.84FRR221 pKa = 11.84QNCDD225 pKa = 3.09GIMKK229 pKa = 9.86SGCLGTIIFNSLLQLAIHH247 pKa = 6.18VLSCMRR253 pKa = 11.84SGEE256 pKa = 4.08DD257 pKa = 3.45LKK259 pKa = 11.14PFYY262 pKa = 10.9AVGDD266 pKa = 3.76DD267 pKa = 4.73TIQEE271 pKa = 4.42TCSEE275 pKa = 4.46EE276 pKa = 3.9YY277 pKa = 9.16WDD279 pKa = 3.94EE280 pKa = 4.15VKK282 pKa = 10.78KK283 pKa = 11.03CGAIVKK289 pKa = 9.2EE290 pKa = 4.9RR291 pKa = 11.84IGGWPAEE298 pKa = 4.15FIGIYY303 pKa = 10.68FNDD306 pKa = 4.0TVGLPMYY313 pKa = 8.17ATKK316 pKa = 10.98NFFRR320 pKa = 11.84LLFAEE325 pKa = 5.4AEE327 pKa = 4.14LLDD330 pKa = 5.71DD331 pKa = 4.27ILAGYY336 pKa = 7.89MRR338 pKa = 11.84LYY340 pKa = 11.05ANSPLFTILNEE351 pKa = 3.62LAIRR355 pKa = 11.84KK356 pKa = 8.39GVTLSKK362 pKa = 10.64DD363 pKa = 3.42DD364 pKa = 3.88CLTWFNGG371 pKa = 3.39
Molecular weight: 41.98 kDa
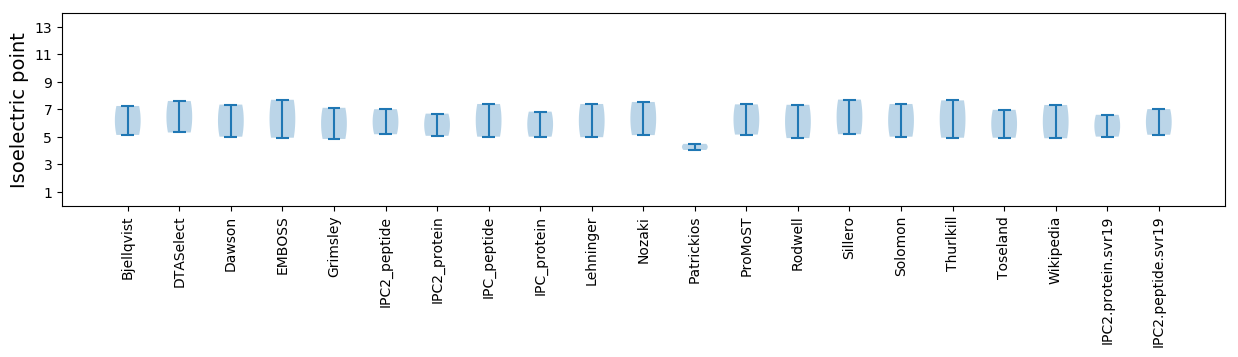
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KFA0|A0A1L3KFA0_9VIRU RNA-directed RNA polymerase OS=Wenling sobemo-like virus 1 OX=1923540 PE=4 SV=1
MM1 pKa = 7.33FGIKK5 pKa = 10.03ALAEE9 pKa = 4.01RR10 pKa = 11.84VLGGVMAVIRR20 pKa = 11.84FVYY23 pKa = 10.0VVGVFALQVAAADD36 pKa = 3.84VLLVAGDD43 pKa = 3.61YY44 pKa = 10.64TNYY47 pKa = 8.8NTVANLANGNGLYY60 pKa = 10.34NWLLPTTYY68 pKa = 10.14FKK70 pKa = 11.03HH71 pKa = 6.53LFTALVVVIAGRR83 pKa = 11.84FMFEE87 pKa = 3.88FRR89 pKa = 11.84STVNALKK96 pKa = 10.33PPSYY100 pKa = 11.0DD101 pKa = 3.19GDD103 pKa = 3.98ADD105 pKa = 3.89RR106 pKa = 11.84MKK108 pKa = 10.83AYY110 pKa = 9.2TFTVKK115 pKa = 10.51DD116 pKa = 3.98FSKK119 pKa = 11.02DD120 pKa = 3.42YY121 pKa = 9.99MRR123 pKa = 11.84QPGFTSEE130 pKa = 3.96SMRR133 pKa = 11.84AGSVIHH139 pKa = 6.4TNLGEE144 pKa = 4.43VKK146 pKa = 10.48GCLHH150 pKa = 6.41VLDD153 pKa = 5.13NNSKK157 pKa = 8.13TVGFAQVVKK166 pKa = 9.77GHH168 pKa = 6.01IVMPEE173 pKa = 4.08HH174 pKa = 7.76VLMAADD180 pKa = 4.75GGRR183 pKa = 11.84LCGPYY188 pKa = 10.34GGLVALNPKK197 pKa = 9.2MFEE200 pKa = 4.04LVKK203 pKa = 10.03TDD205 pKa = 3.49VVVCPLVEE213 pKa = 4.01SHH215 pKa = 6.99RR216 pKa = 11.84KK217 pKa = 8.47ILAVKK222 pKa = 10.23DD223 pKa = 3.21ADD225 pKa = 3.58FRR227 pKa = 11.84PSFRR231 pKa = 11.84NGAFAQMRR239 pKa = 11.84TTPLMDD245 pKa = 5.38PDD247 pKa = 3.34AHH249 pKa = 5.73MVKK252 pKa = 10.02PGCVVQGKK260 pKa = 7.44VQPNDD265 pKa = 3.18MFGMVSFSGSTRR277 pKa = 11.84PGMSGGGYY285 pKa = 9.65YY286 pKa = 10.32VDD288 pKa = 3.92GALVAMHH295 pKa = 6.93LCGGLSNMGVSMGFVMEE312 pKa = 4.41SVARR316 pKa = 11.84PEE318 pKa = 4.49ADD320 pKa = 3.15LDD322 pKa = 3.89TYY324 pKa = 10.46EE325 pKa = 4.36WLMKK329 pKa = 9.88SAEE332 pKa = 4.13YY333 pKa = 10.65EE334 pKa = 5.26DD335 pKa = 4.38GFEE338 pKa = 4.49WDD340 pKa = 3.63NTHH343 pKa = 7.3DD344 pKa = 4.03PSLVIVKK351 pKa = 9.87DD352 pKa = 3.52VNNRR356 pKa = 11.84YY357 pKa = 9.75HH358 pKa = 7.14EE359 pKa = 4.32VDD361 pKa = 3.87LDD363 pKa = 3.68YY364 pKa = 11.51APAEE368 pKa = 4.33FYY370 pKa = 10.95DD371 pKa = 3.75VLGMRR376 pKa = 11.84KK377 pKa = 9.12PKK379 pKa = 10.5VGVRR383 pKa = 11.84NRR385 pKa = 11.84GIDD388 pKa = 3.75YY389 pKa = 10.69SNKK392 pKa = 10.19DD393 pKa = 3.78YY394 pKa = 11.26EE395 pKa = 4.48PEE397 pKa = 3.94AKK399 pKa = 9.88VGYY402 pKa = 9.35DD403 pKa = 3.31HH404 pKa = 7.5QIMKK408 pKa = 9.29TGYY411 pKa = 10.26LKK413 pKa = 10.92DD414 pKa = 3.69PSSLPKK420 pKa = 10.59EE421 pKa = 4.02MLPVGTRR428 pKa = 11.84LRR430 pKa = 11.84AFVEE434 pKa = 4.28EE435 pKa = 4.72EE436 pKa = 4.04DD437 pKa = 5.94VEE439 pKa = 4.86DD440 pKa = 4.65QDD442 pKa = 6.53DD443 pKa = 3.84PVQIDD448 pKa = 3.67DD449 pKa = 3.73AFQRR453 pKa = 11.84ARR455 pKa = 11.84DD456 pKa = 3.7RR457 pKa = 11.84LHH459 pKa = 7.52DD460 pKa = 3.79YY461 pKa = 10.29FKK463 pKa = 11.56NLIAEE468 pKa = 4.7LAADD472 pKa = 3.84VARR475 pKa = 11.84PSKK478 pKa = 10.8EE479 pKa = 3.37EE480 pKa = 3.43AAINVEE486 pKa = 4.04PFLEE490 pKa = 4.21EE491 pKa = 3.47SRR493 pKa = 11.84ALVEE497 pKa = 4.12QKK499 pKa = 11.08GKK501 pKa = 10.05ISRR504 pKa = 11.84RR505 pKa = 11.84LQEE508 pKa = 4.41INTVMHH514 pKa = 6.81GAKK517 pKa = 10.01KK518 pKa = 8.96AQKK521 pKa = 9.72VGKK524 pKa = 7.55TQLLASKK531 pKa = 7.98TQTLEE536 pKa = 3.5EE537 pKa = 3.89MRR539 pKa = 11.84QAKK542 pKa = 8.6EE543 pKa = 3.99MKK545 pKa = 9.67KK546 pKa = 9.88HH547 pKa = 6.48ADD549 pKa = 3.08AATQLLRR556 pKa = 11.84TNDD559 pKa = 3.52LKK561 pKa = 11.22KK562 pKa = 10.16KK563 pKa = 10.3SKK565 pKa = 10.17NARR568 pKa = 11.84QNAKK572 pKa = 10.08RR573 pKa = 11.84KK574 pKa = 8.45EE575 pKa = 4.26QVDD578 pKa = 3.82QMRR581 pKa = 11.84LTQDD585 pKa = 3.18YY586 pKa = 11.2LMEE589 pKa = 4.05QLQQKK594 pKa = 8.23TGQTANDD601 pKa = 3.81LHH603 pKa = 7.23QQMLDD608 pKa = 3.06AMRR611 pKa = 11.84ARR613 pKa = 11.84PSTSQAGFIKK623 pKa = 10.55KK624 pKa = 9.9
MM1 pKa = 7.33FGIKK5 pKa = 10.03ALAEE9 pKa = 4.01RR10 pKa = 11.84VLGGVMAVIRR20 pKa = 11.84FVYY23 pKa = 10.0VVGVFALQVAAADD36 pKa = 3.84VLLVAGDD43 pKa = 3.61YY44 pKa = 10.64TNYY47 pKa = 8.8NTVANLANGNGLYY60 pKa = 10.34NWLLPTTYY68 pKa = 10.14FKK70 pKa = 11.03HH71 pKa = 6.53LFTALVVVIAGRR83 pKa = 11.84FMFEE87 pKa = 3.88FRR89 pKa = 11.84STVNALKK96 pKa = 10.33PPSYY100 pKa = 11.0DD101 pKa = 3.19GDD103 pKa = 3.98ADD105 pKa = 3.89RR106 pKa = 11.84MKK108 pKa = 10.83AYY110 pKa = 9.2TFTVKK115 pKa = 10.51DD116 pKa = 3.98FSKK119 pKa = 11.02DD120 pKa = 3.42YY121 pKa = 9.99MRR123 pKa = 11.84QPGFTSEE130 pKa = 3.96SMRR133 pKa = 11.84AGSVIHH139 pKa = 6.4TNLGEE144 pKa = 4.43VKK146 pKa = 10.48GCLHH150 pKa = 6.41VLDD153 pKa = 5.13NNSKK157 pKa = 8.13TVGFAQVVKK166 pKa = 9.77GHH168 pKa = 6.01IVMPEE173 pKa = 4.08HH174 pKa = 7.76VLMAADD180 pKa = 4.75GGRR183 pKa = 11.84LCGPYY188 pKa = 10.34GGLVALNPKK197 pKa = 9.2MFEE200 pKa = 4.04LVKK203 pKa = 10.03TDD205 pKa = 3.49VVVCPLVEE213 pKa = 4.01SHH215 pKa = 6.99RR216 pKa = 11.84KK217 pKa = 8.47ILAVKK222 pKa = 10.23DD223 pKa = 3.21ADD225 pKa = 3.58FRR227 pKa = 11.84PSFRR231 pKa = 11.84NGAFAQMRR239 pKa = 11.84TTPLMDD245 pKa = 5.38PDD247 pKa = 3.34AHH249 pKa = 5.73MVKK252 pKa = 10.02PGCVVQGKK260 pKa = 7.44VQPNDD265 pKa = 3.18MFGMVSFSGSTRR277 pKa = 11.84PGMSGGGYY285 pKa = 9.65YY286 pKa = 10.32VDD288 pKa = 3.92GALVAMHH295 pKa = 6.93LCGGLSNMGVSMGFVMEE312 pKa = 4.41SVARR316 pKa = 11.84PEE318 pKa = 4.49ADD320 pKa = 3.15LDD322 pKa = 3.89TYY324 pKa = 10.46EE325 pKa = 4.36WLMKK329 pKa = 9.88SAEE332 pKa = 4.13YY333 pKa = 10.65EE334 pKa = 5.26DD335 pKa = 4.38GFEE338 pKa = 4.49WDD340 pKa = 3.63NTHH343 pKa = 7.3DD344 pKa = 4.03PSLVIVKK351 pKa = 9.87DD352 pKa = 3.52VNNRR356 pKa = 11.84YY357 pKa = 9.75HH358 pKa = 7.14EE359 pKa = 4.32VDD361 pKa = 3.87LDD363 pKa = 3.68YY364 pKa = 11.51APAEE368 pKa = 4.33FYY370 pKa = 10.95DD371 pKa = 3.75VLGMRR376 pKa = 11.84KK377 pKa = 9.12PKK379 pKa = 10.5VGVRR383 pKa = 11.84NRR385 pKa = 11.84GIDD388 pKa = 3.75YY389 pKa = 10.69SNKK392 pKa = 10.19DD393 pKa = 3.78YY394 pKa = 11.26EE395 pKa = 4.48PEE397 pKa = 3.94AKK399 pKa = 9.88VGYY402 pKa = 9.35DD403 pKa = 3.31HH404 pKa = 7.5QIMKK408 pKa = 9.29TGYY411 pKa = 10.26LKK413 pKa = 10.92DD414 pKa = 3.69PSSLPKK420 pKa = 10.59EE421 pKa = 4.02MLPVGTRR428 pKa = 11.84LRR430 pKa = 11.84AFVEE434 pKa = 4.28EE435 pKa = 4.72EE436 pKa = 4.04DD437 pKa = 5.94VEE439 pKa = 4.86DD440 pKa = 4.65QDD442 pKa = 6.53DD443 pKa = 3.84PVQIDD448 pKa = 3.67DD449 pKa = 3.73AFQRR453 pKa = 11.84ARR455 pKa = 11.84DD456 pKa = 3.7RR457 pKa = 11.84LHH459 pKa = 7.52DD460 pKa = 3.79YY461 pKa = 10.29FKK463 pKa = 11.56NLIAEE468 pKa = 4.7LAADD472 pKa = 3.84VARR475 pKa = 11.84PSKK478 pKa = 10.8EE479 pKa = 3.37EE480 pKa = 3.43AAINVEE486 pKa = 4.04PFLEE490 pKa = 4.21EE491 pKa = 3.47SRR493 pKa = 11.84ALVEE497 pKa = 4.12QKK499 pKa = 11.08GKK501 pKa = 10.05ISRR504 pKa = 11.84RR505 pKa = 11.84LQEE508 pKa = 4.41INTVMHH514 pKa = 6.81GAKK517 pKa = 10.01KK518 pKa = 8.96AQKK521 pKa = 9.72VGKK524 pKa = 7.55TQLLASKK531 pKa = 7.98TQTLEE536 pKa = 3.5EE537 pKa = 3.89MRR539 pKa = 11.84QAKK542 pKa = 8.6EE543 pKa = 3.99MKK545 pKa = 9.67KK546 pKa = 9.88HH547 pKa = 6.48ADD549 pKa = 3.08AATQLLRR556 pKa = 11.84TNDD559 pKa = 3.52LKK561 pKa = 11.22KK562 pKa = 10.16KK563 pKa = 10.3SKK565 pKa = 10.17NARR568 pKa = 11.84QNAKK572 pKa = 10.08RR573 pKa = 11.84KK574 pKa = 8.45EE575 pKa = 4.26QVDD578 pKa = 3.82QMRR581 pKa = 11.84LTQDD585 pKa = 3.18YY586 pKa = 11.2LMEE589 pKa = 4.05QLQQKK594 pKa = 8.23TGQTANDD601 pKa = 3.81LHH603 pKa = 7.23QQMLDD608 pKa = 3.06AMRR611 pKa = 11.84ARR613 pKa = 11.84PSTSQAGFIKK623 pKa = 10.55KK624 pKa = 9.9
Molecular weight: 69.65 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
995 |
371 |
624 |
497.5 |
55.81 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.638 ± 1.369 | 1.307 ± 0.518 |
7.236 ± 0.189 | 5.729 ± 0.123 |
5.126 ± 0.982 | 7.538 ± 0.17 |
2.111 ± 0.3 | 3.719 ± 1.347 |
6.633 ± 0.592 | 9.045 ± 0.729 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.819 ± 1.013 | 4.121 ± 0.047 |
4.221 ± 0.056 | 3.317 ± 1.199 |
5.226 ± 0.228 | 5.226 ± 0.757 |
4.824 ± 0.181 | 8.543 ± 0.935 |
1.307 ± 0.846 | 3.317 ± 0.214 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
