
Acetobacteraceae bacterium AT-5844
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodospirillales; Acetobacteraceae; unclassified Acetobacteraceae
Average proteome isoelectric point is 6.89
Get precalculated fractions of proteins
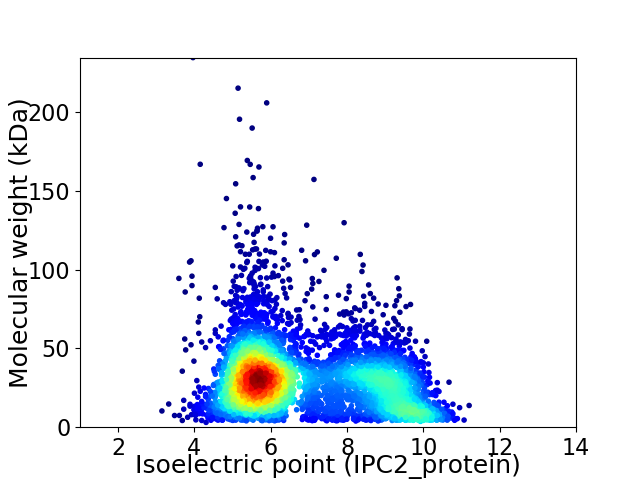
Virtual 2D-PAGE plot for 5288 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|H0A0L9|H0A0L9_9PROT CsaA protein OS=Acetobacteraceae bacterium AT-5844 OX=1054213 GN=HMPREF9946_02360 PE=4 SV=1
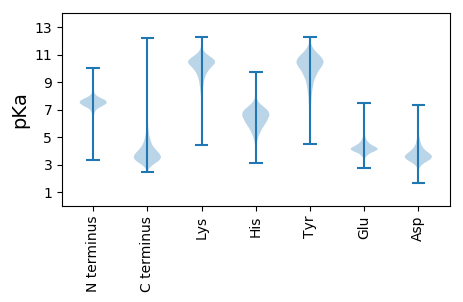
MM1 pKa = 8.14PDD3 pKa = 5.45LITLDD8 pKa = 3.77KK9 pKa = 11.16PEE11 pKa = 4.49DD12 pKa = 4.63AIVLHH17 pKa = 6.9PGDD20 pKa = 4.8NIQEE24 pKa = 4.05AVLAGGEE31 pKa = 4.43GAVFWLEE38 pKa = 3.61AGVYY42 pKa = 9.46RR43 pKa = 11.84MQEE46 pKa = 3.77IEE48 pKa = 4.26PLNGQSFYY56 pKa = 10.89GASGAVLNGSRR67 pKa = 11.84LLTDD71 pKa = 4.0FTQIDD76 pKa = 4.1GHH78 pKa = 5.9WVAEE82 pKa = 4.33GQTQEE87 pKa = 4.04GEE89 pKa = 3.93RR90 pKa = 11.84RR91 pKa = 11.84EE92 pKa = 3.95VDD94 pKa = 3.31EE95 pKa = 4.71AAEE98 pKa = 4.54GAQRR102 pKa = 11.84PGYY105 pKa = 9.25PDD107 pKa = 3.05AVYY110 pKa = 11.39VNDD113 pKa = 3.78QPLKK117 pKa = 10.04PVEE120 pKa = 4.29SLDD123 pKa = 3.93DD124 pKa = 3.93LTSGTYY130 pKa = 10.28YY131 pKa = 10.7FDD133 pKa = 3.88YY134 pKa = 11.26DD135 pKa = 3.11EE136 pKa = 4.66DD137 pKa = 5.01RR138 pKa = 11.84IYY140 pKa = 11.03FGDD143 pKa = 4.23DD144 pKa = 3.2PNGQTVEE151 pKa = 4.18AAVGTHH157 pKa = 7.2AIASTADD164 pKa = 3.26NVTVSGLTVEE174 pKa = 5.61KK175 pKa = 10.57YY176 pKa = 7.68ATPTQFAAITGSGEE190 pKa = 3.55NWIVSNNEE198 pKa = 2.84VRR200 pKa = 11.84LNYY203 pKa = 10.37GVGISVGSDD212 pKa = 3.0GQILNNFVHH221 pKa = 7.73DD222 pKa = 4.27NGQMGLDD229 pKa = 3.21AGGTNILVEE238 pKa = 4.35GNEE241 pKa = 3.96IASNGFWSGIDD252 pKa = 3.6VFWEE256 pKa = 4.39GGGAKK261 pKa = 8.79FTEE264 pKa = 4.27TDD266 pKa = 3.45GLVVRR271 pKa = 11.84NNYY274 pKa = 9.88SHH276 pKa = 7.67DD277 pKa = 3.77NNGYY281 pKa = 9.17GLWTDD286 pKa = 3.02IDD288 pKa = 4.41NINTVYY294 pKa = 10.08EE295 pKa = 4.4GNRR298 pKa = 11.84IEE300 pKa = 4.18YY301 pKa = 10.54NSGGGINHH309 pKa = 7.43EE310 pKa = 3.87ISYY313 pKa = 10.26AASIHH318 pKa = 7.18DD319 pKa = 3.66NTFVGNGGNGLTWLWGSAIQVQNSQDD345 pKa = 3.42VEE347 pKa = 4.1IFNNFIDD354 pKa = 4.47ASSGGNGIGLIQQDD368 pKa = 3.69RR369 pKa = 11.84GEE371 pKa = 4.48GANGPWITVNNNVHH385 pKa = 7.26DD386 pKa = 4.08NTLILSDD393 pKa = 3.69MSGVGAVADD402 pKa = 3.95YY403 pKa = 11.37DD404 pKa = 4.27EE405 pKa = 4.49EE406 pKa = 4.6TMLSGGNVFNNNDD419 pKa = 3.57YY420 pKa = 11.03QVLDD424 pKa = 3.66VEE426 pKa = 4.51NDD428 pKa = 3.06QFAFGEE434 pKa = 4.87FYY436 pKa = 10.72TFEE439 pKa = 5.33DD440 pKa = 3.9FTKK443 pKa = 10.69AANQEE448 pKa = 4.06AGGTLVFAA456 pKa = 5.68
MM1 pKa = 8.14PDD3 pKa = 5.45LITLDD8 pKa = 3.77KK9 pKa = 11.16PEE11 pKa = 4.49DD12 pKa = 4.63AIVLHH17 pKa = 6.9PGDD20 pKa = 4.8NIQEE24 pKa = 4.05AVLAGGEE31 pKa = 4.43GAVFWLEE38 pKa = 3.61AGVYY42 pKa = 9.46RR43 pKa = 11.84MQEE46 pKa = 3.77IEE48 pKa = 4.26PLNGQSFYY56 pKa = 10.89GASGAVLNGSRR67 pKa = 11.84LLTDD71 pKa = 4.0FTQIDD76 pKa = 4.1GHH78 pKa = 5.9WVAEE82 pKa = 4.33GQTQEE87 pKa = 4.04GEE89 pKa = 3.93RR90 pKa = 11.84RR91 pKa = 11.84EE92 pKa = 3.95VDD94 pKa = 3.31EE95 pKa = 4.71AAEE98 pKa = 4.54GAQRR102 pKa = 11.84PGYY105 pKa = 9.25PDD107 pKa = 3.05AVYY110 pKa = 11.39VNDD113 pKa = 3.78QPLKK117 pKa = 10.04PVEE120 pKa = 4.29SLDD123 pKa = 3.93DD124 pKa = 3.93LTSGTYY130 pKa = 10.28YY131 pKa = 10.7FDD133 pKa = 3.88YY134 pKa = 11.26DD135 pKa = 3.11EE136 pKa = 4.66DD137 pKa = 5.01RR138 pKa = 11.84IYY140 pKa = 11.03FGDD143 pKa = 4.23DD144 pKa = 3.2PNGQTVEE151 pKa = 4.18AAVGTHH157 pKa = 7.2AIASTADD164 pKa = 3.26NVTVSGLTVEE174 pKa = 5.61KK175 pKa = 10.57YY176 pKa = 7.68ATPTQFAAITGSGEE190 pKa = 3.55NWIVSNNEE198 pKa = 2.84VRR200 pKa = 11.84LNYY203 pKa = 10.37GVGISVGSDD212 pKa = 3.0GQILNNFVHH221 pKa = 7.73DD222 pKa = 4.27NGQMGLDD229 pKa = 3.21AGGTNILVEE238 pKa = 4.35GNEE241 pKa = 3.96IASNGFWSGIDD252 pKa = 3.6VFWEE256 pKa = 4.39GGGAKK261 pKa = 8.79FTEE264 pKa = 4.27TDD266 pKa = 3.45GLVVRR271 pKa = 11.84NNYY274 pKa = 9.88SHH276 pKa = 7.67DD277 pKa = 3.77NNGYY281 pKa = 9.17GLWTDD286 pKa = 3.02IDD288 pKa = 4.41NINTVYY294 pKa = 10.08EE295 pKa = 4.4GNRR298 pKa = 11.84IEE300 pKa = 4.18YY301 pKa = 10.54NSGGGINHH309 pKa = 7.43EE310 pKa = 3.87ISYY313 pKa = 10.26AASIHH318 pKa = 7.18DD319 pKa = 3.66NTFVGNGGNGLTWLWGSAIQVQNSQDD345 pKa = 3.42VEE347 pKa = 4.1IFNNFIDD354 pKa = 4.47ASSGGNGIGLIQQDD368 pKa = 3.69RR369 pKa = 11.84GEE371 pKa = 4.48GANGPWITVNNNVHH385 pKa = 7.26DD386 pKa = 4.08NTLILSDD393 pKa = 3.69MSGVGAVADD402 pKa = 3.95YY403 pKa = 11.37DD404 pKa = 4.27EE405 pKa = 4.49EE406 pKa = 4.6TMLSGGNVFNNNDD419 pKa = 3.57YY420 pKa = 11.03QVLDD424 pKa = 3.66VEE426 pKa = 4.51NDD428 pKa = 3.06QFAFGEE434 pKa = 4.87FYY436 pKa = 10.72TFEE439 pKa = 5.33DD440 pKa = 3.9FTKK443 pKa = 10.69AANQEE448 pKa = 4.06AGGTLVFAA456 pKa = 5.68
Molecular weight: 49.05 kDa
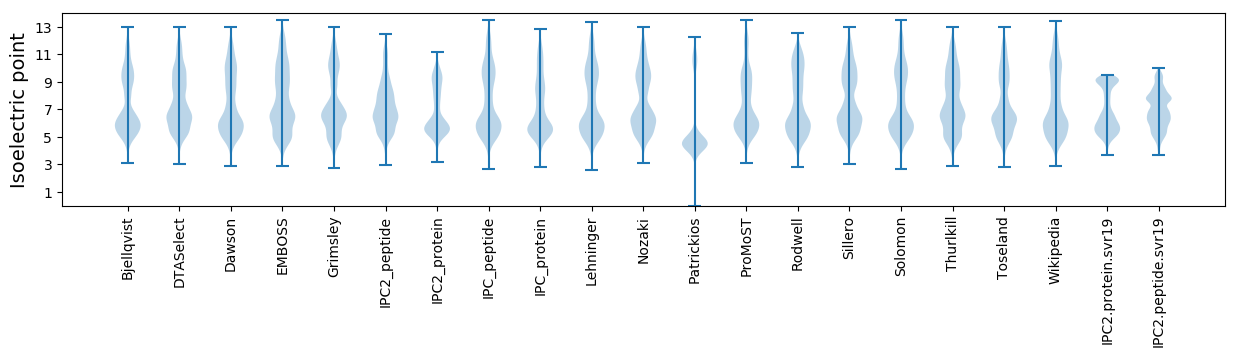
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|H0A5U5|H0A5U5_9PROT Aminotransferase class I/II OS=Acetobacteraceae bacterium AT-5844 OX=1054213 GN=HMPREF9946_04200 PE=4 SV=1
MM1 pKa = 7.27TGIAAHH7 pKa = 6.86IAAAITRR14 pKa = 11.84RR15 pKa = 11.84GARR18 pKa = 11.84SSRR21 pKa = 11.84GRR23 pKa = 11.84RR24 pKa = 11.84GRR26 pKa = 11.84VAARR30 pKa = 11.84LLLRR34 pKa = 11.84LAAGLGLDD42 pKa = 3.68LLTRR46 pKa = 11.84FLLGAGSGFLSGAPLFLSLARR67 pKa = 11.84GLFRR71 pKa = 11.84RR72 pKa = 11.84AAQHH76 pKa = 5.84QRR78 pKa = 11.84LTLTRR83 pKa = 11.84LALTLGFQAAMLLQRR98 pKa = 11.84AQAGRR103 pKa = 11.84LLTLVQGAVAGSVGALRR120 pKa = 11.84ASGRR124 pKa = 11.84ALRR127 pKa = 11.84AGRR130 pKa = 11.84GALL133 pKa = 3.52
MM1 pKa = 7.27TGIAAHH7 pKa = 6.86IAAAITRR14 pKa = 11.84RR15 pKa = 11.84GARR18 pKa = 11.84SSRR21 pKa = 11.84GRR23 pKa = 11.84RR24 pKa = 11.84GRR26 pKa = 11.84VAARR30 pKa = 11.84LLLRR34 pKa = 11.84LAAGLGLDD42 pKa = 3.68LLTRR46 pKa = 11.84FLLGAGSGFLSGAPLFLSLARR67 pKa = 11.84GLFRR71 pKa = 11.84RR72 pKa = 11.84AAQHH76 pKa = 5.84QRR78 pKa = 11.84LTLTRR83 pKa = 11.84LALTLGFQAAMLLQRR98 pKa = 11.84AQAGRR103 pKa = 11.84LLTLVQGAVAGSVGALRR120 pKa = 11.84ASGRR124 pKa = 11.84ALRR127 pKa = 11.84AGRR130 pKa = 11.84GALL133 pKa = 3.52
Molecular weight: 13.78 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1570546 |
28 |
2255 |
297.0 |
31.98 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.849 ± 0.054 | 0.829 ± 0.009 |
4.97 ± 0.026 | 5.664 ± 0.03 |
3.366 ± 0.02 | 9.079 ± 0.035 |
2.03 ± 0.017 | 4.348 ± 0.023 |
2.309 ± 0.025 | 10.936 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.562 ± 0.017 | 2.24 ± 0.02 |
6.005 ± 0.033 | 3.283 ± 0.02 |
7.88 ± 0.032 | 4.989 ± 0.026 |
5.099 ± 0.023 | 7.228 ± 0.026 |
1.467 ± 0.015 | 1.869 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
