
Roseburia sp. CAG:309
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Lachnospiraceae; Roseburia; environmental samples
Average proteome isoelectric point is 6.17
Get precalculated fractions of proteins
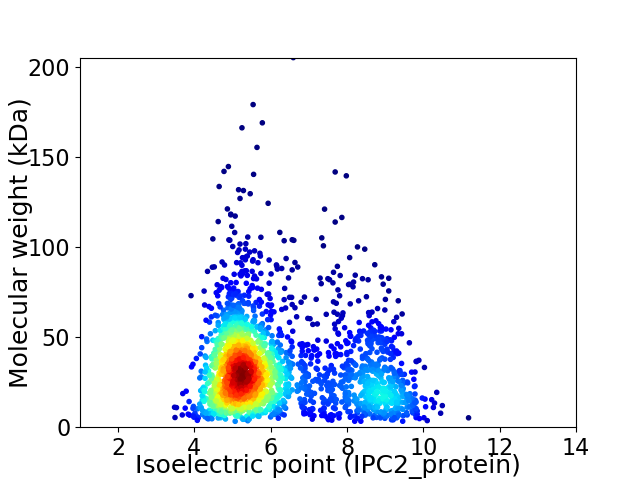
Virtual 2D-PAGE plot for 2223 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
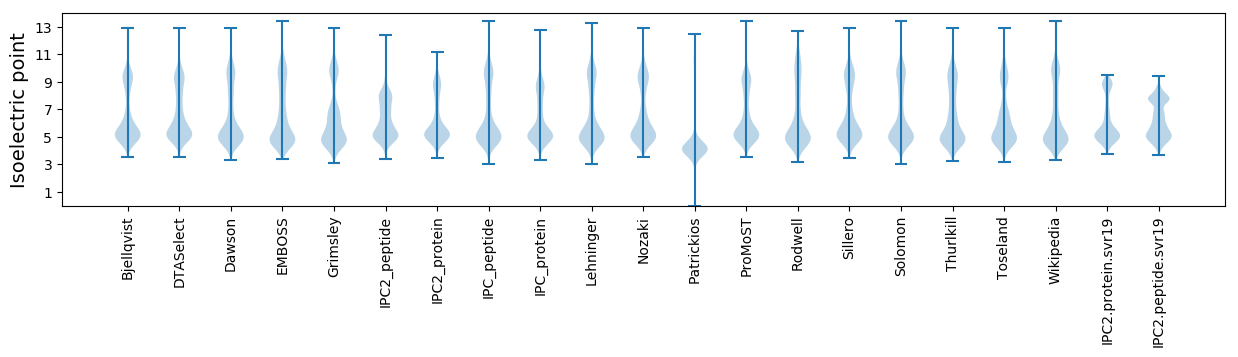
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R6YWA9|R6YWA9_9FIRM Subtilisin-like serine protease OS=Roseburia sp. CAG:309 OX=1262945 GN=BN600_01888 PE=3 SV=1
MM1 pKa = 7.44KK2 pKa = 10.26KK3 pKa = 10.36KK4 pKa = 10.15IVLCIVLAGLIFSLAGCAEE23 pKa = 4.26EE24 pKa = 5.31AGQPEE29 pKa = 4.7AGVQQEE35 pKa = 3.92NSAVSEE41 pKa = 4.34EE42 pKa = 4.28EE43 pKa = 3.98FAGDD47 pKa = 3.83GAWTILVYY55 pKa = 10.99LCGTDD60 pKa = 4.05LEE62 pKa = 4.63SGYY65 pKa = 11.12GAATMDD71 pKa = 4.11LGEE74 pKa = 4.25MLDD77 pKa = 3.99RR78 pKa = 11.84DD79 pKa = 3.9FGSQVRR85 pKa = 11.84FVVQTGGCTQWQNEE99 pKa = 4.85VIPEE103 pKa = 3.88DD104 pKa = 4.25AIARR108 pKa = 11.84YY109 pKa = 9.31EE110 pKa = 4.14IINGDD115 pKa = 3.44MQEE118 pKa = 3.97ISRR121 pKa = 11.84MDD123 pKa = 3.79QANMAEE129 pKa = 4.46STTLQDD135 pKa = 3.69FVTFGMEE142 pKa = 3.72NYY144 pKa = 10.31GNNKK148 pKa = 9.69LGLILWDD155 pKa = 3.55HH156 pKa = 6.68GGGSVAGVCLDD167 pKa = 3.79EE168 pKa = 5.73NYY170 pKa = 9.0GTDD173 pKa = 3.45TLDD176 pKa = 3.63LSEE179 pKa = 4.99IRR181 pKa = 11.84QALDD185 pKa = 4.25GIPRR189 pKa = 11.84KK190 pKa = 9.99FNFIGYY196 pKa = 7.56DD197 pKa = 3.18ACLMGSVEE205 pKa = 4.24NAKK208 pKa = 9.34MLAPFANYY216 pKa = 9.21MIGSEE221 pKa = 4.13EE222 pKa = 4.27LEE224 pKa = 4.47SGGGWDD230 pKa = 3.62YY231 pKa = 11.87AAIGAYY237 pKa = 9.46LQQNPGAPAADD248 pKa = 4.03LGKK251 pKa = 10.08EE252 pKa = 3.96ICDD255 pKa = 3.49SFYY258 pKa = 11.16KK259 pKa = 10.4SCEE262 pKa = 3.83ALGEE266 pKa = 4.16EE267 pKa = 4.63STATLSVTDD276 pKa = 4.04LSKK279 pKa = 10.53IQKK282 pKa = 9.52VCDD285 pKa = 3.55EE286 pKa = 4.66FNTVAEE292 pKa = 4.33TMKK295 pKa = 10.93GSTEE299 pKa = 3.67NMEE302 pKa = 4.53AFGEE306 pKa = 4.18IAKK309 pKa = 10.44GIRR312 pKa = 11.84KK313 pKa = 7.93AQNYY317 pKa = 8.88GGNTPSEE324 pKa = 5.03GYY326 pKa = 11.13SNMVDD331 pKa = 3.49LGDD334 pKa = 3.65MAKK337 pKa = 10.59NIAGTVDD344 pKa = 3.62TSALQAAIAEE354 pKa = 4.16AVIYY358 pKa = 8.27QVKK361 pKa = 10.45GEE363 pKa = 3.98NRR365 pKa = 11.84QNANGLSVYY374 pKa = 10.48YY375 pKa = 9.37PLQIEE380 pKa = 4.5DD381 pKa = 3.96EE382 pKa = 4.58EE383 pKa = 4.21EE384 pKa = 4.0LKK386 pKa = 10.15TFEE389 pKa = 4.8TICVSDD395 pKa = 4.9PYY397 pKa = 10.91TDD399 pKa = 5.4FIGTMVYY406 pKa = 10.38GAANGSIEE414 pKa = 4.38EE415 pKa = 4.15YY416 pKa = 10.53SGEE419 pKa = 4.25NDD421 pKa = 2.7WSAADD426 pKa = 3.61QSGHH430 pKa = 5.88GLGLVTAEE438 pKa = 3.99QNQIQIAEE446 pKa = 4.16EE447 pKa = 3.58QHH449 pKa = 6.76LNDD452 pKa = 4.13DD453 pKa = 4.14GYY455 pKa = 9.69YY456 pKa = 8.88TFTIAEE462 pKa = 4.01EE463 pKa = 4.13SLPNVASVQYY473 pKa = 10.2NLYY476 pKa = 10.04MKK478 pKa = 10.56FDD480 pKa = 4.68DD481 pKa = 4.38EE482 pKa = 4.75TPLVYY487 pKa = 10.55LGSDD491 pKa = 2.57NWLDD495 pKa = 3.72YY496 pKa = 11.33DD497 pKa = 4.04DD498 pKa = 3.9QTGVVTDD505 pKa = 3.91TFQGYY510 pKa = 7.73WPALPDD516 pKa = 3.77GSLLSMYY523 pKa = 10.15VVEE526 pKa = 4.48EE527 pKa = 4.42SEE529 pKa = 4.98DD530 pKa = 3.6YY531 pKa = 10.99TIFSSPIVLNGRR543 pKa = 11.84EE544 pKa = 4.1TNLRR548 pKa = 11.84FAYY551 pKa = 10.28LYY553 pKa = 11.23GEE555 pKa = 4.34TEE557 pKa = 5.62DD558 pKa = 3.98DD559 pKa = 4.8GEE561 pKa = 4.23WLVLGAWDD569 pKa = 6.14GIDD572 pKa = 4.77DD573 pKa = 5.97DD574 pKa = 5.51SGQADD579 pKa = 3.43KK580 pKa = 11.42EE581 pKa = 4.03MTEE584 pKa = 3.87IQKK587 pKa = 10.96GDD589 pKa = 4.07VIRR592 pKa = 11.84PIYY595 pKa = 10.36VVVDD599 pKa = 3.52QEE601 pKa = 4.71SGDD604 pKa = 3.6TTEE607 pKa = 4.81IKK609 pKa = 10.27GDD611 pKa = 3.31KK612 pKa = 10.56YY613 pKa = 10.61RR614 pKa = 11.84VKK616 pKa = 10.84KK617 pKa = 10.41NFNITEE623 pKa = 4.19PKK625 pKa = 10.28LPEE628 pKa = 3.89GEE630 pKa = 4.4YY631 pKa = 10.46YY632 pKa = 10.84YY633 pKa = 11.43SFGIDD638 pKa = 3.7DD639 pKa = 5.25LYY641 pKa = 11.67GSTKK645 pKa = 10.6YY646 pKa = 10.82LDD648 pKa = 5.57LINFWVNKK656 pKa = 8.42KK657 pKa = 10.67GKK659 pKa = 10.29VEE661 pKa = 3.95LSYY664 pKa = 11.68
MM1 pKa = 7.44KK2 pKa = 10.26KK3 pKa = 10.36KK4 pKa = 10.15IVLCIVLAGLIFSLAGCAEE23 pKa = 4.26EE24 pKa = 5.31AGQPEE29 pKa = 4.7AGVQQEE35 pKa = 3.92NSAVSEE41 pKa = 4.34EE42 pKa = 4.28EE43 pKa = 3.98FAGDD47 pKa = 3.83GAWTILVYY55 pKa = 10.99LCGTDD60 pKa = 4.05LEE62 pKa = 4.63SGYY65 pKa = 11.12GAATMDD71 pKa = 4.11LGEE74 pKa = 4.25MLDD77 pKa = 3.99RR78 pKa = 11.84DD79 pKa = 3.9FGSQVRR85 pKa = 11.84FVVQTGGCTQWQNEE99 pKa = 4.85VIPEE103 pKa = 3.88DD104 pKa = 4.25AIARR108 pKa = 11.84YY109 pKa = 9.31EE110 pKa = 4.14IINGDD115 pKa = 3.44MQEE118 pKa = 3.97ISRR121 pKa = 11.84MDD123 pKa = 3.79QANMAEE129 pKa = 4.46STTLQDD135 pKa = 3.69FVTFGMEE142 pKa = 3.72NYY144 pKa = 10.31GNNKK148 pKa = 9.69LGLILWDD155 pKa = 3.55HH156 pKa = 6.68GGGSVAGVCLDD167 pKa = 3.79EE168 pKa = 5.73NYY170 pKa = 9.0GTDD173 pKa = 3.45TLDD176 pKa = 3.63LSEE179 pKa = 4.99IRR181 pKa = 11.84QALDD185 pKa = 4.25GIPRR189 pKa = 11.84KK190 pKa = 9.99FNFIGYY196 pKa = 7.56DD197 pKa = 3.18ACLMGSVEE205 pKa = 4.24NAKK208 pKa = 9.34MLAPFANYY216 pKa = 9.21MIGSEE221 pKa = 4.13EE222 pKa = 4.27LEE224 pKa = 4.47SGGGWDD230 pKa = 3.62YY231 pKa = 11.87AAIGAYY237 pKa = 9.46LQQNPGAPAADD248 pKa = 4.03LGKK251 pKa = 10.08EE252 pKa = 3.96ICDD255 pKa = 3.49SFYY258 pKa = 11.16KK259 pKa = 10.4SCEE262 pKa = 3.83ALGEE266 pKa = 4.16EE267 pKa = 4.63STATLSVTDD276 pKa = 4.04LSKK279 pKa = 10.53IQKK282 pKa = 9.52VCDD285 pKa = 3.55EE286 pKa = 4.66FNTVAEE292 pKa = 4.33TMKK295 pKa = 10.93GSTEE299 pKa = 3.67NMEE302 pKa = 4.53AFGEE306 pKa = 4.18IAKK309 pKa = 10.44GIRR312 pKa = 11.84KK313 pKa = 7.93AQNYY317 pKa = 8.88GGNTPSEE324 pKa = 5.03GYY326 pKa = 11.13SNMVDD331 pKa = 3.49LGDD334 pKa = 3.65MAKK337 pKa = 10.59NIAGTVDD344 pKa = 3.62TSALQAAIAEE354 pKa = 4.16AVIYY358 pKa = 8.27QVKK361 pKa = 10.45GEE363 pKa = 3.98NRR365 pKa = 11.84QNANGLSVYY374 pKa = 10.48YY375 pKa = 9.37PLQIEE380 pKa = 4.5DD381 pKa = 3.96EE382 pKa = 4.58EE383 pKa = 4.21EE384 pKa = 4.0LKK386 pKa = 10.15TFEE389 pKa = 4.8TICVSDD395 pKa = 4.9PYY397 pKa = 10.91TDD399 pKa = 5.4FIGTMVYY406 pKa = 10.38GAANGSIEE414 pKa = 4.38EE415 pKa = 4.15YY416 pKa = 10.53SGEE419 pKa = 4.25NDD421 pKa = 2.7WSAADD426 pKa = 3.61QSGHH430 pKa = 5.88GLGLVTAEE438 pKa = 3.99QNQIQIAEE446 pKa = 4.16EE447 pKa = 3.58QHH449 pKa = 6.76LNDD452 pKa = 4.13DD453 pKa = 4.14GYY455 pKa = 9.69YY456 pKa = 8.88TFTIAEE462 pKa = 4.01EE463 pKa = 4.13SLPNVASVQYY473 pKa = 10.2NLYY476 pKa = 10.04MKK478 pKa = 10.56FDD480 pKa = 4.68DD481 pKa = 4.38EE482 pKa = 4.75TPLVYY487 pKa = 10.55LGSDD491 pKa = 2.57NWLDD495 pKa = 3.72YY496 pKa = 11.33DD497 pKa = 4.04DD498 pKa = 3.9QTGVVTDD505 pKa = 3.91TFQGYY510 pKa = 7.73WPALPDD516 pKa = 3.77GSLLSMYY523 pKa = 10.15VVEE526 pKa = 4.48EE527 pKa = 4.42SEE529 pKa = 4.98DD530 pKa = 3.6YY531 pKa = 10.99TIFSSPIVLNGRR543 pKa = 11.84EE544 pKa = 4.1TNLRR548 pKa = 11.84FAYY551 pKa = 10.28LYY553 pKa = 11.23GEE555 pKa = 4.34TEE557 pKa = 5.62DD558 pKa = 3.98DD559 pKa = 4.8GEE561 pKa = 4.23WLVLGAWDD569 pKa = 6.14GIDD572 pKa = 4.77DD573 pKa = 5.97DD574 pKa = 5.51SGQADD579 pKa = 3.43KK580 pKa = 11.42EE581 pKa = 4.03MTEE584 pKa = 3.87IQKK587 pKa = 10.96GDD589 pKa = 4.07VIRR592 pKa = 11.84PIYY595 pKa = 10.36VVVDD599 pKa = 3.52QEE601 pKa = 4.71SGDD604 pKa = 3.6TTEE607 pKa = 4.81IKK609 pKa = 10.27GDD611 pKa = 3.31KK612 pKa = 10.56YY613 pKa = 10.61RR614 pKa = 11.84VKK616 pKa = 10.84KK617 pKa = 10.41NFNITEE623 pKa = 4.19PKK625 pKa = 10.28LPEE628 pKa = 3.89GEE630 pKa = 4.4YY631 pKa = 10.46YY632 pKa = 10.84YY633 pKa = 11.43SFGIDD638 pKa = 3.7DD639 pKa = 5.25LYY641 pKa = 11.67GSTKK645 pKa = 10.6YY646 pKa = 10.82LDD648 pKa = 5.57LINFWVNKK656 pKa = 8.42KK657 pKa = 10.67GKK659 pKa = 10.29VEE661 pKa = 3.95LSYY664 pKa = 11.68
Molecular weight: 72.95 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R6YFX9|R6YFX9_9FIRM Uncharacterized protein OS=Roseburia sp. CAG:309 OX=1262945 GN=BN600_01208 PE=4 SV=1
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 8.86KK9 pKa = 8.02KK10 pKa = 9.86SRR12 pKa = 11.84SRR14 pKa = 11.84VHH16 pKa = 6.67GFRR19 pKa = 11.84KK20 pKa = 10.01RR21 pKa = 11.84MSTSNGRR28 pKa = 11.84KK29 pKa = 8.05VLSRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.2GRR39 pKa = 11.84KK40 pKa = 8.79KK41 pKa = 10.63LSAA44 pKa = 3.95
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 8.86KK9 pKa = 8.02KK10 pKa = 9.86SRR12 pKa = 11.84SRR14 pKa = 11.84VHH16 pKa = 6.67GFRR19 pKa = 11.84KK20 pKa = 10.01RR21 pKa = 11.84MSTSNGRR28 pKa = 11.84KK29 pKa = 8.05VLSRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.2GRR39 pKa = 11.84KK40 pKa = 8.79KK41 pKa = 10.63LSAA44 pKa = 3.95
Molecular weight: 5.2 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
679977 |
29 |
1929 |
305.9 |
34.39 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.301 ± 0.05 | 1.542 ± 0.022 |
5.519 ± 0.039 | 7.823 ± 0.066 |
4.104 ± 0.041 | 6.887 ± 0.048 |
1.722 ± 0.024 | 7.21 ± 0.043 |
7.65 ± 0.051 | 8.589 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.105 ± 0.027 | 4.262 ± 0.033 |
3.258 ± 0.031 | 3.334 ± 0.03 |
4.249 ± 0.038 | 5.666 ± 0.042 |
5.596 ± 0.048 | 7.119 ± 0.044 |
0.89 ± 0.021 | 4.169 ± 0.041 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
