
Rotavirus B (isolate RVB/Human/China/ADRV/1982) (RV-B) (Rotavirus B (isolate adult diarrhea rotavirus))
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Resentoviricetes; Reovirales; Reoviridae; Sedoreovirinae; Rotavirus; Rotavirus B
Average proteome isoelectric point is 6.41
Get precalculated fractions of proteins
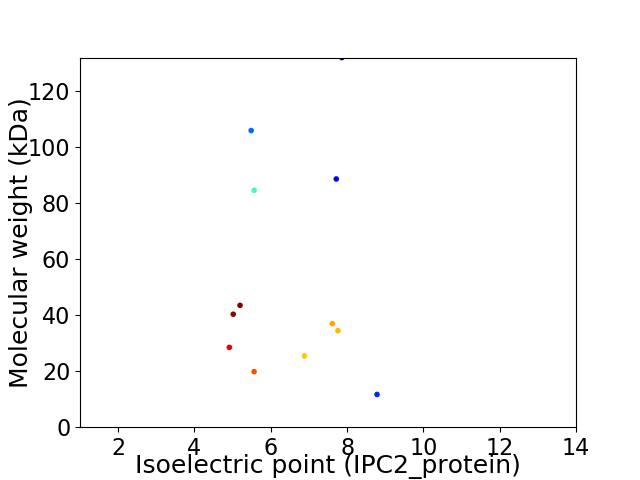
Virtual 2D-PAGE plot for 12 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
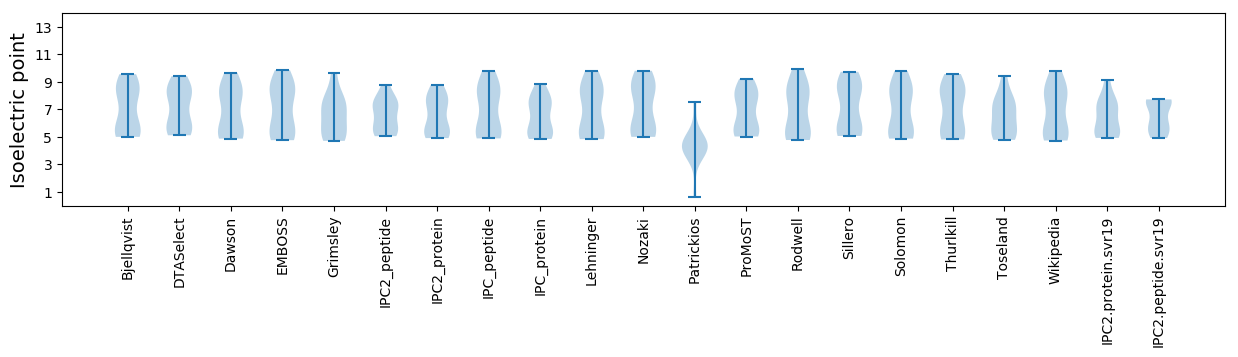
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D6NGG0|D6NGG0_ROTGA Nonstructural protein NSP1-1 OS=Rotavirus B (isolate RVB/Human/China/ADRV/1982) OX=10942 PE=4 SV=1
MM1 pKa = 7.78ASLPLLVLAAAVNAQLNIVPSVHH24 pKa = 6.89PEE26 pKa = 3.86VCVLYY31 pKa = 10.78ADD33 pKa = 5.01DD34 pKa = 4.87HH35 pKa = 7.2QSDD38 pKa = 3.83ANKK41 pKa = 10.23FNGNFTQIFHH51 pKa = 7.04SYY53 pKa = 11.01NSITLSFMSYY63 pKa = 10.24SSSSYY68 pKa = 11.12DD69 pKa = 3.35VIDD72 pKa = 3.17IMSRR76 pKa = 11.84YY77 pKa = 9.48DD78 pKa = 3.86LSSCSILAIDD88 pKa = 4.74VFDD91 pKa = 4.98ASMDD95 pKa = 3.83FNVFLQSTNNCSKK108 pKa = 10.78YY109 pKa = 9.86NANKK113 pKa = 9.08IHH115 pKa = 6.37YY116 pKa = 8.63IKK118 pKa = 10.57LPRR121 pKa = 11.84GEE123 pKa = 4.29EE124 pKa = 3.76WFSYY128 pKa = 10.32SKK130 pKa = 10.84NLKK133 pKa = 9.63FCPLSDD139 pKa = 3.85SLIGMYY145 pKa = 10.34CDD147 pKa = 3.57TQLSDD152 pKa = 3.69TYY154 pKa = 11.31FEE156 pKa = 4.61ISTGGTYY163 pKa = 10.29EE164 pKa = 4.04VTDD167 pKa = 3.7VPEE170 pKa = 4.39FTQMGYY176 pKa = 7.7TFHH179 pKa = 8.12SSEE182 pKa = 5.04DD183 pKa = 3.61FYY185 pKa = 11.53LCHH188 pKa = 7.18RR189 pKa = 11.84ISSEE193 pKa = 3.11AWLNYY198 pKa = 9.38HH199 pKa = 6.99LFYY202 pKa = 10.62RR203 pKa = 11.84DD204 pKa = 3.31YY205 pKa = 10.9DD206 pKa = 3.83VSGVISKK213 pKa = 9.89QINWGNVWSGFKK225 pKa = 10.1TFAQVLYY232 pKa = 10.46KK233 pKa = 10.41ILDD236 pKa = 3.49LFFNSKK242 pKa = 10.65RR243 pKa = 11.84NVEE246 pKa = 4.02PRR248 pKa = 11.84AA249 pKa = 3.51
MM1 pKa = 7.78ASLPLLVLAAAVNAQLNIVPSVHH24 pKa = 6.89PEE26 pKa = 3.86VCVLYY31 pKa = 10.78ADD33 pKa = 5.01DD34 pKa = 4.87HH35 pKa = 7.2QSDD38 pKa = 3.83ANKK41 pKa = 10.23FNGNFTQIFHH51 pKa = 7.04SYY53 pKa = 11.01NSITLSFMSYY63 pKa = 10.24SSSSYY68 pKa = 11.12DD69 pKa = 3.35VIDD72 pKa = 3.17IMSRR76 pKa = 11.84YY77 pKa = 9.48DD78 pKa = 3.86LSSCSILAIDD88 pKa = 4.74VFDD91 pKa = 4.98ASMDD95 pKa = 3.83FNVFLQSTNNCSKK108 pKa = 10.78YY109 pKa = 9.86NANKK113 pKa = 9.08IHH115 pKa = 6.37YY116 pKa = 8.63IKK118 pKa = 10.57LPRR121 pKa = 11.84GEE123 pKa = 4.29EE124 pKa = 3.76WFSYY128 pKa = 10.32SKK130 pKa = 10.84NLKK133 pKa = 9.63FCPLSDD139 pKa = 3.85SLIGMYY145 pKa = 10.34CDD147 pKa = 3.57TQLSDD152 pKa = 3.69TYY154 pKa = 11.31FEE156 pKa = 4.61ISTGGTYY163 pKa = 10.29EE164 pKa = 4.04VTDD167 pKa = 3.7VPEE170 pKa = 4.39FTQMGYY176 pKa = 7.7TFHH179 pKa = 8.12SSEE182 pKa = 5.04DD183 pKa = 3.61FYY185 pKa = 11.53LCHH188 pKa = 7.18RR189 pKa = 11.84ISSEE193 pKa = 3.11AWLNYY198 pKa = 9.38HH199 pKa = 6.99LFYY202 pKa = 10.62RR203 pKa = 11.84DD204 pKa = 3.31YY205 pKa = 10.9DD206 pKa = 3.83VSGVISKK213 pKa = 9.89QINWGNVWSGFKK225 pKa = 10.1TFAQVLYY232 pKa = 10.46KK233 pKa = 10.41ILDD236 pKa = 3.49LFFNSKK242 pKa = 10.65RR243 pKa = 11.84NVEE246 pKa = 4.02PRR248 pKa = 11.84AA249 pKa = 3.51
Molecular weight: 28.53 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D6NGG1|D6NGG1_ROTGA Nonstructural protein NSP1-2 OS=Rotavirus B (isolate RVB/Human/China/ADRV/1982) OX=10942 PE=4 SV=1
MM1 pKa = 7.84GNRR4 pKa = 11.84QSSAQLNSHH13 pKa = 6.25LTHH16 pKa = 7.03INSQNSNLFISDD28 pKa = 3.04SKK30 pKa = 9.88TAVFHH35 pKa = 5.6TQHH38 pKa = 6.61ILLAAGVGIIATLLVLLLCSCVLNCYY64 pKa = 10.22LCRR67 pKa = 11.84KK68 pKa = 9.37LKK70 pKa = 9.25RR71 pKa = 11.84TNGVSSLLEE80 pKa = 3.64RR81 pKa = 11.84NIRR84 pKa = 11.84QNGSSAKK91 pKa = 9.89IYY93 pKa = 9.41VKK95 pKa = 10.32PVMQSSTIIEE105 pKa = 4.17EE106 pKa = 4.13AA107 pKa = 3.36
MM1 pKa = 7.84GNRR4 pKa = 11.84QSSAQLNSHH13 pKa = 6.25LTHH16 pKa = 7.03INSQNSNLFISDD28 pKa = 3.04SKK30 pKa = 9.88TAVFHH35 pKa = 5.6TQHH38 pKa = 6.61ILLAAGVGIIATLLVLLLCSCVLNCYY64 pKa = 10.22LCRR67 pKa = 11.84KK68 pKa = 9.37LKK70 pKa = 9.25RR71 pKa = 11.84TNGVSSLLEE80 pKa = 3.64RR81 pKa = 11.84NIRR84 pKa = 11.84QNGSSAKK91 pKa = 9.89IYY93 pKa = 9.41VKK95 pKa = 10.32PVMQSSTIIEE105 pKa = 4.17EE106 pKa = 4.13AA107 pKa = 3.36
Molecular weight: 11.7 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5712 |
107 |
1160 |
476.0 |
54.38 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.777 ± 0.388 | 1.366 ± 0.238 |
6.548 ± 0.306 | 5.41 ± 0.756 |
4.482 ± 0.454 | 3.957 ± 0.505 |
2.223 ± 0.219 | 7.668 ± 0.326 |
6.285 ± 0.506 | 8.841 ± 0.588 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.819 ± 0.271 | 6.057 ± 0.445 |
3.589 ± 0.355 | 4.132 ± 0.271 |
5.182 ± 0.387 | 8.298 ± 0.486 |
6.46 ± 0.29 | 5.83 ± 0.422 |
0.963 ± 0.124 | 4.114 ± 0.484 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
