
Azobacteroides phage ProJPt-Bp1
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Podoviridae; unclassified Podoviridae; crAss-like viruses
Average proteome isoelectric point is 6.15
Get precalculated fractions of proteins
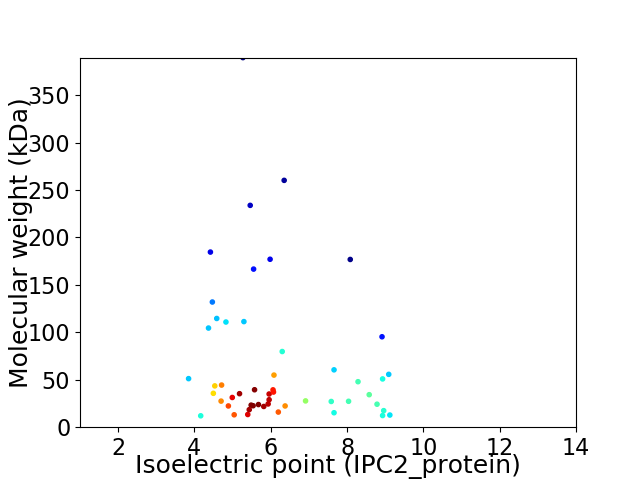
Virtual 2D-PAGE plot for 52 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1V1FGX9|A0A1V1FGX9_9CAUD VIP2 Actin-ADP-ribosylating toxin family protein OS=Azobacteroides phage ProJPt-Bp1 OX=1920526 PE=4 SV=1
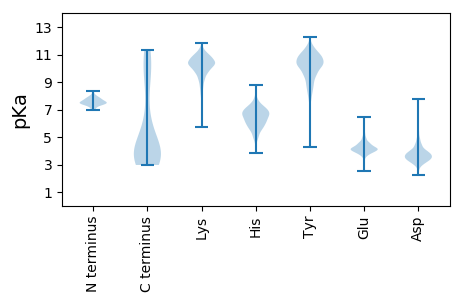
MM1 pKa = 7.26AQHH4 pKa = 6.8IDD6 pKa = 3.34MYY8 pKa = 10.29DD9 pKa = 3.06IRR11 pKa = 11.84IGPDD15 pKa = 2.63KK16 pKa = 10.82FIIIDD21 pKa = 3.43NHH23 pKa = 5.9ALYY26 pKa = 8.04TTFSIEE32 pKa = 3.86FEE34 pKa = 4.04IEE36 pKa = 3.23EE37 pKa = 5.05GYY39 pKa = 10.08IQFTDD44 pKa = 3.78DD45 pKa = 4.07LVGDD49 pKa = 3.97VYY51 pKa = 10.78IGQIANMTIDD61 pKa = 3.74GVAAATTGEE70 pKa = 4.18EE71 pKa = 4.69LADD74 pKa = 3.63QLGDD78 pKa = 3.56YY79 pKa = 7.56MTDD82 pKa = 3.07VLSEE86 pKa = 4.08ADD88 pKa = 3.81IEE90 pKa = 4.67TSGATQYY97 pKa = 11.78DD98 pKa = 4.0FVDD101 pKa = 4.66DD102 pKa = 4.08GLGFQVKK109 pKa = 10.01HH110 pKa = 6.12EE111 pKa = 4.12NSVVYY116 pKa = 10.17DD117 pKa = 3.7YY118 pKa = 11.52SANKK122 pKa = 9.22LVPVTSVSTTTGTGRR137 pKa = 11.84INNLTTGIMISHH149 pKa = 7.66TGTSYY154 pKa = 10.68IPSLTIHH161 pKa = 6.31SGDD164 pKa = 3.43PAMLFVFKK172 pKa = 10.83NGTSTLVALSSDD184 pKa = 3.38GTLATTKK191 pKa = 9.97SQNYY195 pKa = 6.93NTMPNGEE202 pKa = 4.57YY203 pKa = 8.73VTKK206 pKa = 10.82ALADD210 pKa = 3.5TAYY213 pKa = 10.73VDD215 pKa = 3.85TFASLVGGSFTVTRR229 pKa = 11.84RR230 pKa = 11.84GNIIFNYY237 pKa = 10.03SAPQWEE243 pKa = 4.58YY244 pKa = 10.77TFNNLSGGFVVAKK257 pKa = 9.36NGTVIYY263 pKa = 8.78TYY265 pKa = 9.66TPPVVPDD272 pKa = 3.05QDD274 pKa = 3.79VYY276 pKa = 11.56TFADD280 pKa = 3.89YY281 pKa = 7.6PTTSGFQVSKK291 pKa = 10.81NSTPIYY297 pKa = 9.46TYY299 pKa = 10.98SAVLDD304 pKa = 4.05TFTDD308 pKa = 3.42EE309 pKa = 5.35DD310 pKa = 4.4DD311 pKa = 3.94GFKK314 pKa = 10.43VARR317 pKa = 11.84GGSTIYY323 pKa = 10.24TYY325 pKa = 10.16TPPDD329 pKa = 3.47SAVDD333 pKa = 3.71TFSDD337 pKa = 4.0NDD339 pKa = 3.7DD340 pKa = 4.64DD341 pKa = 4.47GFKK344 pKa = 9.59VTRR347 pKa = 11.84DD348 pKa = 3.21GQVIYY353 pKa = 10.63NYY355 pKa = 10.12DD356 pKa = 3.56PPIVGGSLDD365 pKa = 4.11FEE367 pKa = 4.88TTTDD371 pKa = 2.81QYY373 pKa = 11.94GFVTGFEE380 pKa = 4.43VKK382 pKa = 10.62DD383 pKa = 3.75EE384 pKa = 4.21EE385 pKa = 5.53DD386 pKa = 3.57EE387 pKa = 4.37VLYY390 pKa = 10.11TYY392 pKa = 11.67ADD394 pKa = 3.47AQYY397 pKa = 10.54GAQTIGTAMNLYY409 pKa = 10.52KK410 pKa = 10.43SGQLVYY416 pKa = 9.7TYY418 pKa = 10.7DD419 pKa = 3.13AAAIVGSGFARR430 pKa = 11.84SIIKK434 pKa = 10.1SDD436 pKa = 3.6SGDD439 pKa = 3.65EE440 pKa = 3.97EE441 pKa = 5.8LMISQIADD449 pKa = 3.39EE450 pKa = 4.44VGGYY454 pKa = 8.61TMALLQSSTIGGGGKK469 pKa = 8.23RR470 pKa = 11.84TITT473 pKa = 3.49
MM1 pKa = 7.26AQHH4 pKa = 6.8IDD6 pKa = 3.34MYY8 pKa = 10.29DD9 pKa = 3.06IRR11 pKa = 11.84IGPDD15 pKa = 2.63KK16 pKa = 10.82FIIIDD21 pKa = 3.43NHH23 pKa = 5.9ALYY26 pKa = 8.04TTFSIEE32 pKa = 3.86FEE34 pKa = 4.04IEE36 pKa = 3.23EE37 pKa = 5.05GYY39 pKa = 10.08IQFTDD44 pKa = 3.78DD45 pKa = 4.07LVGDD49 pKa = 3.97VYY51 pKa = 10.78IGQIANMTIDD61 pKa = 3.74GVAAATTGEE70 pKa = 4.18EE71 pKa = 4.69LADD74 pKa = 3.63QLGDD78 pKa = 3.56YY79 pKa = 7.56MTDD82 pKa = 3.07VLSEE86 pKa = 4.08ADD88 pKa = 3.81IEE90 pKa = 4.67TSGATQYY97 pKa = 11.78DD98 pKa = 4.0FVDD101 pKa = 4.66DD102 pKa = 4.08GLGFQVKK109 pKa = 10.01HH110 pKa = 6.12EE111 pKa = 4.12NSVVYY116 pKa = 10.17DD117 pKa = 3.7YY118 pKa = 11.52SANKK122 pKa = 9.22LVPVTSVSTTTGTGRR137 pKa = 11.84INNLTTGIMISHH149 pKa = 7.66TGTSYY154 pKa = 10.68IPSLTIHH161 pKa = 6.31SGDD164 pKa = 3.43PAMLFVFKK172 pKa = 10.83NGTSTLVALSSDD184 pKa = 3.38GTLATTKK191 pKa = 9.97SQNYY195 pKa = 6.93NTMPNGEE202 pKa = 4.57YY203 pKa = 8.73VTKK206 pKa = 10.82ALADD210 pKa = 3.5TAYY213 pKa = 10.73VDD215 pKa = 3.85TFASLVGGSFTVTRR229 pKa = 11.84RR230 pKa = 11.84GNIIFNYY237 pKa = 10.03SAPQWEE243 pKa = 4.58YY244 pKa = 10.77TFNNLSGGFVVAKK257 pKa = 9.36NGTVIYY263 pKa = 8.78TYY265 pKa = 9.66TPPVVPDD272 pKa = 3.05QDD274 pKa = 3.79VYY276 pKa = 11.56TFADD280 pKa = 3.89YY281 pKa = 7.6PTTSGFQVSKK291 pKa = 10.81NSTPIYY297 pKa = 9.46TYY299 pKa = 10.98SAVLDD304 pKa = 4.05TFTDD308 pKa = 3.42EE309 pKa = 5.35DD310 pKa = 4.4DD311 pKa = 3.94GFKK314 pKa = 10.43VARR317 pKa = 11.84GGSTIYY323 pKa = 10.24TYY325 pKa = 10.16TPPDD329 pKa = 3.47SAVDD333 pKa = 3.71TFSDD337 pKa = 4.0NDD339 pKa = 3.7DD340 pKa = 4.64DD341 pKa = 4.47GFKK344 pKa = 9.59VTRR347 pKa = 11.84DD348 pKa = 3.21GQVIYY353 pKa = 10.63NYY355 pKa = 10.12DD356 pKa = 3.56PPIVGGSLDD365 pKa = 4.11FEE367 pKa = 4.88TTTDD371 pKa = 2.81QYY373 pKa = 11.94GFVTGFEE380 pKa = 4.43VKK382 pKa = 10.62DD383 pKa = 3.75EE384 pKa = 4.21EE385 pKa = 5.53DD386 pKa = 3.57EE387 pKa = 4.37VLYY390 pKa = 10.11TYY392 pKa = 11.67ADD394 pKa = 3.47AQYY397 pKa = 10.54GAQTIGTAMNLYY409 pKa = 10.52KK410 pKa = 10.43SGQLVYY416 pKa = 9.7TYY418 pKa = 10.7DD419 pKa = 3.13AAAIVGSGFARR430 pKa = 11.84SIIKK434 pKa = 10.1SDD436 pKa = 3.6SGDD439 pKa = 3.65EE440 pKa = 3.97EE441 pKa = 5.8LMISQIADD449 pKa = 3.39EE450 pKa = 4.44VGGYY454 pKa = 8.61TMALLQSSTIGGGGKK469 pKa = 8.23RR470 pKa = 11.84TITT473 pKa = 3.49
Molecular weight: 51.2 kDa
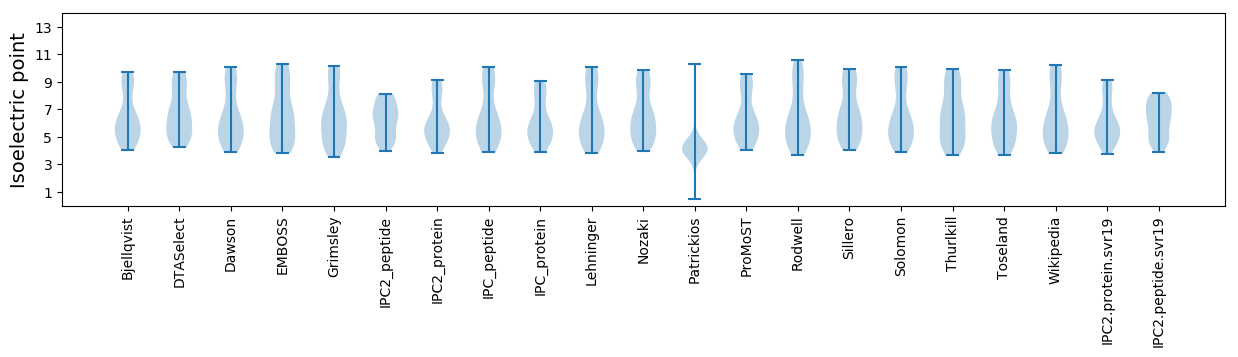
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1V1FY48|A0A1V1FY48_9CAUD Phage protein inside capsid D OS=Azobacteroides phage ProJPt-Bp1 OX=1920526 PE=4 SV=1
MM1 pKa = 7.48KK2 pKa = 10.1PFYY5 pKa = 10.76NYY7 pKa = 9.12TDD9 pKa = 3.43PRR11 pKa = 11.84DD12 pKa = 3.44NRR14 pKa = 11.84FVFKK18 pKa = 10.67GSEE21 pKa = 3.89NRR23 pKa = 11.84PSAVAMEE30 pKa = 5.13DD31 pKa = 2.93IVRR34 pKa = 11.84GAKK37 pKa = 9.98NNPTFIDD44 pKa = 4.05FQEE47 pKa = 4.09RR48 pKa = 11.84QYY50 pKa = 11.42PGTKK54 pKa = 9.3KK55 pKa = 10.52AFRR58 pKa = 11.84KK59 pKa = 8.42WLAYY63 pKa = 10.17DD64 pKa = 3.85PQRR67 pKa = 11.84MIDD70 pKa = 3.61PATNMPYY77 pKa = 10.95DD78 pKa = 4.01LAAWNVPTEE87 pKa = 4.34MPSDD91 pKa = 3.74TPIDD95 pKa = 3.92FLSPTAKK102 pKa = 10.32HH103 pKa = 5.99GGSVQRR109 pKa = 11.84FAHH112 pKa = 6.24GGSISGLDD120 pKa = 3.4GFKK123 pKa = 11.05YY124 pKa = 10.24PIRR127 pKa = 11.84RR128 pKa = 11.84YY129 pKa = 8.86ATGGSAAVPASGTLEE144 pKa = 4.48AIAAASAAADD154 pKa = 3.45KK155 pKa = 9.91ATAKK159 pKa = 10.31GGSWGDD165 pKa = 3.69FANTAAQTVNSSGVSISDD183 pKa = 3.57MIRR186 pKa = 11.84NLFRR190 pKa = 11.84KK191 pKa = 9.92RR192 pKa = 11.84PTVGANDD199 pKa = 3.76YY200 pKa = 9.2YY201 pKa = 10.54YY202 pKa = 11.22KK203 pKa = 11.07VMEE206 pKa = 4.15EE207 pKa = 3.99ALRR210 pKa = 11.84GSLAKK215 pKa = 10.35AQSGRR220 pKa = 11.84GADD223 pKa = 4.07LLPPTKK229 pKa = 10.1PQDD232 pKa = 3.21IEE234 pKa = 4.31AEE236 pKa = 4.02PDD238 pKa = 2.84IDD240 pKa = 3.81PAYY243 pKa = 7.31MTSEE247 pKa = 4.41RR248 pKa = 11.84NNMTGVVAKK257 pKa = 10.94ALMDD261 pKa = 4.54KK262 pKa = 10.8IKK264 pKa = 10.37DD265 pKa = 3.66TDD267 pKa = 3.86SEE269 pKa = 4.51RR270 pKa = 11.84SDD272 pKa = 3.33ALEE275 pKa = 3.93RR276 pKa = 11.84LASRR280 pKa = 11.84RR281 pKa = 11.84RR282 pKa = 11.84EE283 pKa = 3.78AGRR286 pKa = 11.84AGDD289 pKa = 3.68APTRR293 pKa = 11.84LAMILEE299 pKa = 4.39AQRR302 pKa = 11.84QYY304 pKa = 11.16GNEE307 pKa = 4.01MKK309 pKa = 10.55GARR312 pKa = 11.84AAADD316 pKa = 3.21ASNRR320 pKa = 11.84DD321 pKa = 3.12TRR323 pKa = 11.84AEE325 pKa = 4.12FARR328 pKa = 11.84TMAAIGDD335 pKa = 4.07ADD337 pKa = 3.63SAKK340 pKa = 10.75AMEE343 pKa = 4.53ASRR346 pKa = 11.84HH347 pKa = 5.13NIEE350 pKa = 3.91EE351 pKa = 4.14KK352 pKa = 10.46TRR354 pKa = 11.84VDD356 pKa = 3.4DD357 pKa = 4.41LNARR361 pKa = 11.84RR362 pKa = 11.84HH363 pKa = 4.95AAHH366 pKa = 6.4RR367 pKa = 11.84QARR370 pKa = 11.84SAQNYY375 pKa = 9.73AYY377 pKa = 8.93EE378 pKa = 4.06QEE380 pKa = 4.2KK381 pKa = 10.56RR382 pKa = 11.84AFNNQQRR389 pKa = 11.84MDD391 pKa = 3.42EE392 pKa = 4.21YY393 pKa = 11.33NRR395 pKa = 11.84SKK397 pKa = 11.01EE398 pKa = 3.92SDD400 pKa = 3.2ALRR403 pKa = 11.84LQLAEE408 pKa = 3.66LHH410 pKa = 5.79GRR412 pKa = 11.84RR413 pKa = 11.84GQEE416 pKa = 3.15ADD418 pKa = 3.73LAKK421 pKa = 10.81EE422 pKa = 3.77MAKK425 pKa = 10.23EE426 pKa = 3.92NRR428 pKa = 11.84WKK430 pKa = 10.39SWRR433 pKa = 11.84TWLKK437 pKa = 10.45LLGMGAGATIGGMTGGVAGAGLGLSLGSVLKK468 pKa = 10.7HH469 pKa = 5.6GGSVRR474 pKa = 11.84RR475 pKa = 11.84YY476 pKa = 10.37SKK478 pKa = 11.35GGMQNAWMNQGKK490 pKa = 10.19DD491 pKa = 2.47IDD493 pKa = 4.31LYY495 pKa = 11.72VRR497 pKa = 11.84TKK499 pKa = 10.19KK500 pKa = 10.15QRR502 pKa = 11.84NPFRR506 pKa = 5.79
MM1 pKa = 7.48KK2 pKa = 10.1PFYY5 pKa = 10.76NYY7 pKa = 9.12TDD9 pKa = 3.43PRR11 pKa = 11.84DD12 pKa = 3.44NRR14 pKa = 11.84FVFKK18 pKa = 10.67GSEE21 pKa = 3.89NRR23 pKa = 11.84PSAVAMEE30 pKa = 5.13DD31 pKa = 2.93IVRR34 pKa = 11.84GAKK37 pKa = 9.98NNPTFIDD44 pKa = 4.05FQEE47 pKa = 4.09RR48 pKa = 11.84QYY50 pKa = 11.42PGTKK54 pKa = 9.3KK55 pKa = 10.52AFRR58 pKa = 11.84KK59 pKa = 8.42WLAYY63 pKa = 10.17DD64 pKa = 3.85PQRR67 pKa = 11.84MIDD70 pKa = 3.61PATNMPYY77 pKa = 10.95DD78 pKa = 4.01LAAWNVPTEE87 pKa = 4.34MPSDD91 pKa = 3.74TPIDD95 pKa = 3.92FLSPTAKK102 pKa = 10.32HH103 pKa = 5.99GGSVQRR109 pKa = 11.84FAHH112 pKa = 6.24GGSISGLDD120 pKa = 3.4GFKK123 pKa = 11.05YY124 pKa = 10.24PIRR127 pKa = 11.84RR128 pKa = 11.84YY129 pKa = 8.86ATGGSAAVPASGTLEE144 pKa = 4.48AIAAASAAADD154 pKa = 3.45KK155 pKa = 9.91ATAKK159 pKa = 10.31GGSWGDD165 pKa = 3.69FANTAAQTVNSSGVSISDD183 pKa = 3.57MIRR186 pKa = 11.84NLFRR190 pKa = 11.84KK191 pKa = 9.92RR192 pKa = 11.84PTVGANDD199 pKa = 3.76YY200 pKa = 9.2YY201 pKa = 10.54YY202 pKa = 11.22KK203 pKa = 11.07VMEE206 pKa = 4.15EE207 pKa = 3.99ALRR210 pKa = 11.84GSLAKK215 pKa = 10.35AQSGRR220 pKa = 11.84GADD223 pKa = 4.07LLPPTKK229 pKa = 10.1PQDD232 pKa = 3.21IEE234 pKa = 4.31AEE236 pKa = 4.02PDD238 pKa = 2.84IDD240 pKa = 3.81PAYY243 pKa = 7.31MTSEE247 pKa = 4.41RR248 pKa = 11.84NNMTGVVAKK257 pKa = 10.94ALMDD261 pKa = 4.54KK262 pKa = 10.8IKK264 pKa = 10.37DD265 pKa = 3.66TDD267 pKa = 3.86SEE269 pKa = 4.51RR270 pKa = 11.84SDD272 pKa = 3.33ALEE275 pKa = 3.93RR276 pKa = 11.84LASRR280 pKa = 11.84RR281 pKa = 11.84RR282 pKa = 11.84EE283 pKa = 3.78AGRR286 pKa = 11.84AGDD289 pKa = 3.68APTRR293 pKa = 11.84LAMILEE299 pKa = 4.39AQRR302 pKa = 11.84QYY304 pKa = 11.16GNEE307 pKa = 4.01MKK309 pKa = 10.55GARR312 pKa = 11.84AAADD316 pKa = 3.21ASNRR320 pKa = 11.84DD321 pKa = 3.12TRR323 pKa = 11.84AEE325 pKa = 4.12FARR328 pKa = 11.84TMAAIGDD335 pKa = 4.07ADD337 pKa = 3.63SAKK340 pKa = 10.75AMEE343 pKa = 4.53ASRR346 pKa = 11.84HH347 pKa = 5.13NIEE350 pKa = 3.91EE351 pKa = 4.14KK352 pKa = 10.46TRR354 pKa = 11.84VDD356 pKa = 3.4DD357 pKa = 4.41LNARR361 pKa = 11.84RR362 pKa = 11.84HH363 pKa = 4.95AAHH366 pKa = 6.4RR367 pKa = 11.84QARR370 pKa = 11.84SAQNYY375 pKa = 9.73AYY377 pKa = 8.93EE378 pKa = 4.06QEE380 pKa = 4.2KK381 pKa = 10.56RR382 pKa = 11.84AFNNQQRR389 pKa = 11.84MDD391 pKa = 3.42EE392 pKa = 4.21YY393 pKa = 11.33NRR395 pKa = 11.84SKK397 pKa = 11.01EE398 pKa = 3.92SDD400 pKa = 3.2ALRR403 pKa = 11.84LQLAEE408 pKa = 3.66LHH410 pKa = 5.79GRR412 pKa = 11.84RR413 pKa = 11.84GQEE416 pKa = 3.15ADD418 pKa = 3.73LAKK421 pKa = 10.81EE422 pKa = 3.77MAKK425 pKa = 10.23EE426 pKa = 3.92NRR428 pKa = 11.84WKK430 pKa = 10.39SWRR433 pKa = 11.84TWLKK437 pKa = 10.45LLGMGAGATIGGMTGGVAGAGLGLSLGSVLKK468 pKa = 10.7HH469 pKa = 5.6GGSVRR474 pKa = 11.84RR475 pKa = 11.84YY476 pKa = 10.37SKK478 pKa = 11.35GGMQNAWMNQGKK490 pKa = 10.19DD491 pKa = 2.47IDD493 pKa = 4.31LYY495 pKa = 11.72VRR497 pKa = 11.84TKK499 pKa = 10.19KK500 pKa = 10.15QRR502 pKa = 11.84NPFRR506 pKa = 5.79
Molecular weight: 55.71 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
31065 |
102 |
3481 |
597.4 |
67.09 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.207 ± 0.336 | 0.592 ± 0.127 |
6.992 ± 0.202 | 6.103 ± 0.243 |
3.827 ± 0.18 | 7.574 ± 0.399 |
2.002 ± 0.12 | 5.765 ± 0.307 |
6.193 ± 0.34 | 6.921 ± 0.285 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.129 ± 0.147 | 5.521 ± 0.166 |
5.453 ± 0.424 | 4.214 ± 0.389 |
4.948 ± 0.291 | 5.981 ± 0.251 |
6.158 ± 0.379 | 5.592 ± 0.289 |
1.336 ± 0.068 | 4.491 ± 0.156 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
