
Beihai tombus-like virus 18
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.58
Get precalculated fractions of proteins
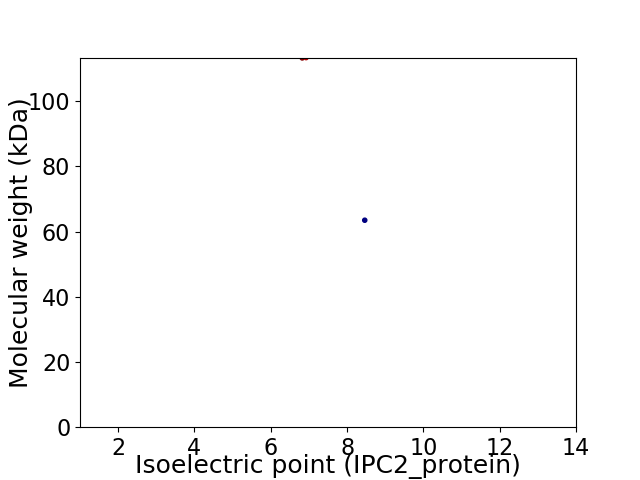
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
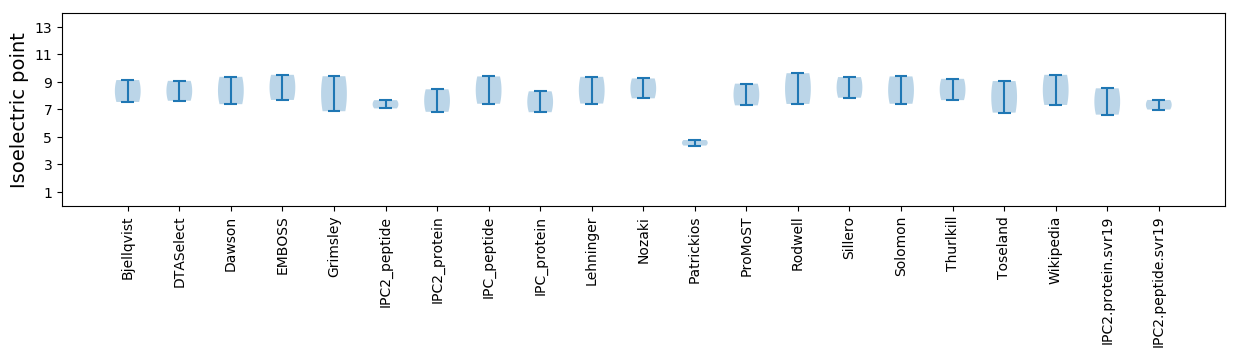
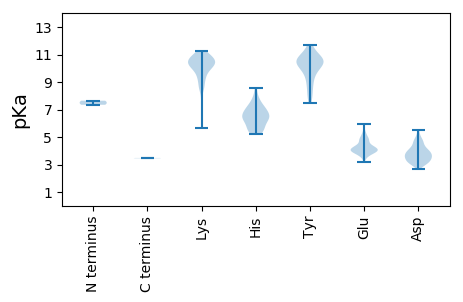
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KFP7|A0A1L3KFP7_9VIRU Uncharacterized protein OS=Beihai tombus-like virus 18 OX=1922721 PE=4 SV=1
MM1 pKa = 7.35PTNKK5 pKa = 10.11GNTKK9 pKa = 9.79KK10 pKa = 10.99ANGNANMKK18 pKa = 10.79KK19 pKa = 9.65MMAKK23 pKa = 9.95SSSKK27 pKa = 10.19SGPRR31 pKa = 11.84SGITPKK37 pKa = 10.66VNLGKK42 pKa = 10.33QKK44 pKa = 11.09SSVKK48 pKa = 9.39WGNDD52 pKa = 2.71RR53 pKa = 11.84CSVSGEE59 pKa = 4.21DD60 pKa = 5.51FIHH63 pKa = 6.12TLQLNEE69 pKa = 4.79LPNSRR74 pKa = 11.84NFAGAVLFPQYY85 pKa = 10.79VNPWRR90 pKa = 11.84LPGTRR95 pKa = 11.84LTQFANLFQRR105 pKa = 11.84FKK107 pKa = 10.27FTRR110 pKa = 11.84FTVKK114 pKa = 10.55YY115 pKa = 8.88IPAVPATVAGQIIMCTDD132 pKa = 3.66TDD134 pKa = 4.04ATWAPIGDD142 pKa = 4.05TQDD145 pKa = 3.5VIRR148 pKa = 11.84MMMAHH153 pKa = 7.9KK154 pKa = 10.23DD155 pKa = 3.07RR156 pKa = 11.84IMFHH160 pKa = 6.76VFDD163 pKa = 4.2NAKK166 pKa = 10.35AVIPGTSKK174 pKa = 10.9PEE176 pKa = 3.99YY177 pKa = 9.06MCDD180 pKa = 3.31AQGQDD185 pKa = 3.2VRR187 pKa = 11.84VNNQGVFWAALVSPVVKK204 pKa = 10.34QDD206 pKa = 3.03GAEE209 pKa = 3.76WTGAVGSFVLEE220 pKa = 3.94WEE222 pKa = 4.79VIFTAARR229 pKa = 11.84ILTPQRR235 pKa = 11.84ITSQSTAKK243 pKa = 10.52AQWTGDD249 pKa = 3.64DD250 pKa = 4.72DD251 pKa = 4.76YY252 pKa = 11.69GASLAQVTGAEE263 pKa = 4.81PNTQTCVAIFRR274 pKa = 11.84DD275 pKa = 3.7VGQAIGMTSGTPYY288 pKa = 10.8FLGSRR293 pKa = 11.84PLSTRR298 pKa = 11.84VVPSEE303 pKa = 3.7YY304 pKa = 9.71EE305 pKa = 3.42RR306 pKa = 11.84AYY308 pKa = 10.97NVYY311 pKa = 8.58GTLRR315 pKa = 11.84GAQQLDD321 pKa = 3.87KK322 pKa = 11.29LDD324 pKa = 5.36LIPVEE329 pKa = 5.06GILTAVDD336 pKa = 4.75FIWWPIGPEE345 pKa = 3.53HH346 pKa = 6.45GTRR349 pKa = 11.84NSTIDD354 pKa = 3.78AKK356 pKa = 11.15VQMSGLSNTTDD367 pKa = 3.03TGLLNVDD374 pKa = 4.45YY375 pKa = 11.21NNTDD379 pKa = 3.13PRR381 pKa = 11.84RR382 pKa = 11.84QADD385 pKa = 4.12EE386 pKa = 4.36YY387 pKa = 10.83FVRR390 pKa = 11.84ATGALPEE397 pKa = 5.35DD398 pKa = 4.28NADD401 pKa = 5.09DD402 pKa = 4.54PSDD405 pKa = 4.02LNGVALIYY413 pKa = 10.43TDD415 pKa = 4.0EE416 pKa = 4.68NDD418 pKa = 3.08VTRR421 pKa = 11.84TVEE424 pKa = 3.89QNQMLIYY431 pKa = 10.6RR432 pKa = 11.84KK433 pKa = 5.68TTPRR437 pKa = 11.84ALATAMGWTDD447 pKa = 3.24VQFQNIYY454 pKa = 10.23GFSVNEE460 pKa = 3.85KK461 pKa = 10.08AFVGVLITALGHH473 pKa = 5.37FVRR476 pKa = 11.84AVSILAKK483 pKa = 9.75AARR486 pKa = 11.84IALAIIRR493 pKa = 11.84EE494 pKa = 4.25VKK496 pKa = 10.0EE497 pKa = 3.91YY498 pKa = 10.5EE499 pKa = 3.94SEE501 pKa = 3.96FFKK504 pKa = 10.92RR505 pKa = 11.84INFSAASEE513 pKa = 4.2LEE515 pKa = 4.21RR516 pKa = 11.84ANGLHH521 pKa = 6.42SGHH524 pKa = 6.34TLSLVKK530 pKa = 10.62NPPRR534 pKa = 11.84QIPSGGFRR542 pKa = 11.84RR543 pKa = 11.84EE544 pKa = 3.73KK545 pKa = 10.6ASNVXILGTGDD556 pKa = 3.54STSLQLVEE564 pKa = 4.72PCPEE568 pKa = 4.2PGVFVFRR575 pKa = 11.84EE576 pKa = 4.25VNDD579 pKa = 3.46EE580 pKa = 4.04HH581 pKa = 7.71QGNLSEE587 pKa = 5.22PDD589 pKa = 3.83DD590 pKa = 4.71SDD592 pKa = 4.24SEE594 pKa = 4.96SIASSDD600 pKa = 3.44PLNEE604 pKa = 3.77HH605 pKa = 6.14FVFRR609 pKa = 11.84RR610 pKa = 11.84NDD612 pKa = 3.41DD613 pKa = 3.11VCYY616 pKa = 10.44VVVNIVNNYY625 pKa = 9.04IFEE628 pKa = 5.3CDD630 pKa = 3.1THH632 pKa = 8.54FDD634 pKa = 4.08FPPGSVWVPDD644 pKa = 3.37LTVFGIEE651 pKa = 4.16PNPGPVCHH659 pKa = 6.62NCGKK663 pKa = 9.81NGHH666 pKa = 6.32IARR669 pKa = 11.84EE670 pKa = 4.1CHH672 pKa = 5.5APKK675 pKa = 10.5AKK677 pKa = 10.26RR678 pKa = 11.84EE679 pKa = 3.96TRR681 pKa = 11.84KK682 pKa = 7.73TVNKK686 pKa = 10.19NLGPRR691 pKa = 11.84RR692 pKa = 11.84DD693 pKa = 3.97GQNKK697 pKa = 8.64LAASVRR703 pKa = 11.84EE704 pKa = 3.96GLAAVAGASDD714 pKa = 3.54AAKK717 pKa = 10.43EE718 pKa = 3.92KK719 pKa = 10.69DD720 pKa = 3.26AEE722 pKa = 4.81AIDD725 pKa = 3.77RR726 pKa = 11.84LKK728 pKa = 10.97EE729 pKa = 3.96VIKK732 pKa = 10.65EE733 pKa = 4.13VVKK736 pKa = 8.59TQKK739 pKa = 10.73DD740 pKa = 3.7EE741 pKa = 4.15EE742 pKa = 4.29EE743 pKa = 4.9KK744 pKa = 10.79DD745 pKa = 3.6VKK747 pKa = 10.29TYY749 pKa = 10.58TPLPGNFTSVIEE761 pKa = 4.38HH762 pKa = 7.14KK763 pKa = 11.15YY764 pKa = 10.11FDD766 pKa = 4.5HH767 pKa = 7.63PDD769 pKa = 4.73FIPTYY774 pKa = 9.87KK775 pKa = 9.08VTSPVRR781 pKa = 11.84ILYY784 pKa = 7.63HH785 pKa = 5.2TLILTLKK792 pKa = 8.74LCKK795 pKa = 10.06YY796 pKa = 9.87YY797 pKa = 10.06PLFGSAVVYY806 pKa = 10.6LLLSLLMKK814 pKa = 10.29RR815 pKa = 11.84INSIMPTGPAFMKK828 pKa = 10.09PISSLFPLMAKK839 pKa = 10.26CFVTLPATFWSLGFSYY855 pKa = 10.53GFAFTYY861 pKa = 9.68VLRR864 pKa = 11.84HH865 pKa = 5.33LKK867 pKa = 10.21FRR869 pKa = 11.84TTYY872 pKa = 8.05EE873 pKa = 4.43TYY875 pKa = 10.73CKK877 pKa = 10.46VLYY880 pKa = 10.46QEE882 pKa = 5.59AEE884 pKa = 3.99GEE886 pKa = 4.12MRR888 pKa = 11.84PEE890 pKa = 4.02RR891 pKa = 11.84MHH893 pKa = 6.34HH894 pKa = 6.1QDD896 pKa = 3.83LIKK899 pKa = 11.06YY900 pKa = 9.47SDD902 pKa = 3.58PKK904 pKa = 8.45TVKK907 pKa = 10.67VKK909 pKa = 9.66VTDD912 pKa = 4.05KK913 pKa = 11.11LICQDD918 pKa = 4.58DD919 pKa = 4.05LFKK922 pKa = 10.35IDD924 pKa = 4.0EE925 pKa = 4.38EE926 pKa = 4.49VVEE929 pKa = 4.28YY930 pKa = 10.44EE931 pKa = 5.04RR932 pKa = 11.84KK933 pKa = 9.26RR934 pKa = 11.84QLNVSLEE941 pKa = 4.15LFSQFSGYY949 pKa = 8.12TMITAPTKK957 pKa = 9.33EE958 pKa = 4.49FYY960 pKa = 10.01MGLQRR965 pKa = 11.84DD966 pKa = 3.81IKK968 pKa = 8.6RR969 pKa = 11.84TCSIGFDD976 pKa = 3.34RR977 pKa = 11.84MDD979 pKa = 3.22MARR982 pKa = 11.84RR983 pKa = 11.84NIFNDD988 pKa = 2.94TATYY992 pKa = 7.46ITAWKK997 pKa = 9.43LSEE1000 pKa = 3.75MTKK1003 pKa = 10.04RR1004 pKa = 11.84DD1005 pKa = 3.42SHH1007 pKa = 5.43EE1008 pKa = 3.9QPLNN1012 pKa = 3.45
MM1 pKa = 7.35PTNKK5 pKa = 10.11GNTKK9 pKa = 9.79KK10 pKa = 10.99ANGNANMKK18 pKa = 10.79KK19 pKa = 9.65MMAKK23 pKa = 9.95SSSKK27 pKa = 10.19SGPRR31 pKa = 11.84SGITPKK37 pKa = 10.66VNLGKK42 pKa = 10.33QKK44 pKa = 11.09SSVKK48 pKa = 9.39WGNDD52 pKa = 2.71RR53 pKa = 11.84CSVSGEE59 pKa = 4.21DD60 pKa = 5.51FIHH63 pKa = 6.12TLQLNEE69 pKa = 4.79LPNSRR74 pKa = 11.84NFAGAVLFPQYY85 pKa = 10.79VNPWRR90 pKa = 11.84LPGTRR95 pKa = 11.84LTQFANLFQRR105 pKa = 11.84FKK107 pKa = 10.27FTRR110 pKa = 11.84FTVKK114 pKa = 10.55YY115 pKa = 8.88IPAVPATVAGQIIMCTDD132 pKa = 3.66TDD134 pKa = 4.04ATWAPIGDD142 pKa = 4.05TQDD145 pKa = 3.5VIRR148 pKa = 11.84MMMAHH153 pKa = 7.9KK154 pKa = 10.23DD155 pKa = 3.07RR156 pKa = 11.84IMFHH160 pKa = 6.76VFDD163 pKa = 4.2NAKK166 pKa = 10.35AVIPGTSKK174 pKa = 10.9PEE176 pKa = 3.99YY177 pKa = 9.06MCDD180 pKa = 3.31AQGQDD185 pKa = 3.2VRR187 pKa = 11.84VNNQGVFWAALVSPVVKK204 pKa = 10.34QDD206 pKa = 3.03GAEE209 pKa = 3.76WTGAVGSFVLEE220 pKa = 3.94WEE222 pKa = 4.79VIFTAARR229 pKa = 11.84ILTPQRR235 pKa = 11.84ITSQSTAKK243 pKa = 10.52AQWTGDD249 pKa = 3.64DD250 pKa = 4.72DD251 pKa = 4.76YY252 pKa = 11.69GASLAQVTGAEE263 pKa = 4.81PNTQTCVAIFRR274 pKa = 11.84DD275 pKa = 3.7VGQAIGMTSGTPYY288 pKa = 10.8FLGSRR293 pKa = 11.84PLSTRR298 pKa = 11.84VVPSEE303 pKa = 3.7YY304 pKa = 9.71EE305 pKa = 3.42RR306 pKa = 11.84AYY308 pKa = 10.97NVYY311 pKa = 8.58GTLRR315 pKa = 11.84GAQQLDD321 pKa = 3.87KK322 pKa = 11.29LDD324 pKa = 5.36LIPVEE329 pKa = 5.06GILTAVDD336 pKa = 4.75FIWWPIGPEE345 pKa = 3.53HH346 pKa = 6.45GTRR349 pKa = 11.84NSTIDD354 pKa = 3.78AKK356 pKa = 11.15VQMSGLSNTTDD367 pKa = 3.03TGLLNVDD374 pKa = 4.45YY375 pKa = 11.21NNTDD379 pKa = 3.13PRR381 pKa = 11.84RR382 pKa = 11.84QADD385 pKa = 4.12EE386 pKa = 4.36YY387 pKa = 10.83FVRR390 pKa = 11.84ATGALPEE397 pKa = 5.35DD398 pKa = 4.28NADD401 pKa = 5.09DD402 pKa = 4.54PSDD405 pKa = 4.02LNGVALIYY413 pKa = 10.43TDD415 pKa = 4.0EE416 pKa = 4.68NDD418 pKa = 3.08VTRR421 pKa = 11.84TVEE424 pKa = 3.89QNQMLIYY431 pKa = 10.6RR432 pKa = 11.84KK433 pKa = 5.68TTPRR437 pKa = 11.84ALATAMGWTDD447 pKa = 3.24VQFQNIYY454 pKa = 10.23GFSVNEE460 pKa = 3.85KK461 pKa = 10.08AFVGVLITALGHH473 pKa = 5.37FVRR476 pKa = 11.84AVSILAKK483 pKa = 9.75AARR486 pKa = 11.84IALAIIRR493 pKa = 11.84EE494 pKa = 4.25VKK496 pKa = 10.0EE497 pKa = 3.91YY498 pKa = 10.5EE499 pKa = 3.94SEE501 pKa = 3.96FFKK504 pKa = 10.92RR505 pKa = 11.84INFSAASEE513 pKa = 4.2LEE515 pKa = 4.21RR516 pKa = 11.84ANGLHH521 pKa = 6.42SGHH524 pKa = 6.34TLSLVKK530 pKa = 10.62NPPRR534 pKa = 11.84QIPSGGFRR542 pKa = 11.84RR543 pKa = 11.84EE544 pKa = 3.73KK545 pKa = 10.6ASNVXILGTGDD556 pKa = 3.54STSLQLVEE564 pKa = 4.72PCPEE568 pKa = 4.2PGVFVFRR575 pKa = 11.84EE576 pKa = 4.25VNDD579 pKa = 3.46EE580 pKa = 4.04HH581 pKa = 7.71QGNLSEE587 pKa = 5.22PDD589 pKa = 3.83DD590 pKa = 4.71SDD592 pKa = 4.24SEE594 pKa = 4.96SIASSDD600 pKa = 3.44PLNEE604 pKa = 3.77HH605 pKa = 6.14FVFRR609 pKa = 11.84RR610 pKa = 11.84NDD612 pKa = 3.41DD613 pKa = 3.11VCYY616 pKa = 10.44VVVNIVNNYY625 pKa = 9.04IFEE628 pKa = 5.3CDD630 pKa = 3.1THH632 pKa = 8.54FDD634 pKa = 4.08FPPGSVWVPDD644 pKa = 3.37LTVFGIEE651 pKa = 4.16PNPGPVCHH659 pKa = 6.62NCGKK663 pKa = 9.81NGHH666 pKa = 6.32IARR669 pKa = 11.84EE670 pKa = 4.1CHH672 pKa = 5.5APKK675 pKa = 10.5AKK677 pKa = 10.26RR678 pKa = 11.84EE679 pKa = 3.96TRR681 pKa = 11.84KK682 pKa = 7.73TVNKK686 pKa = 10.19NLGPRR691 pKa = 11.84RR692 pKa = 11.84DD693 pKa = 3.97GQNKK697 pKa = 8.64LAASVRR703 pKa = 11.84EE704 pKa = 3.96GLAAVAGASDD714 pKa = 3.54AAKK717 pKa = 10.43EE718 pKa = 3.92KK719 pKa = 10.69DD720 pKa = 3.26AEE722 pKa = 4.81AIDD725 pKa = 3.77RR726 pKa = 11.84LKK728 pKa = 10.97EE729 pKa = 3.96VIKK732 pKa = 10.65EE733 pKa = 4.13VVKK736 pKa = 8.59TQKK739 pKa = 10.73DD740 pKa = 3.7EE741 pKa = 4.15EE742 pKa = 4.29EE743 pKa = 4.9KK744 pKa = 10.79DD745 pKa = 3.6VKK747 pKa = 10.29TYY749 pKa = 10.58TPLPGNFTSVIEE761 pKa = 4.38HH762 pKa = 7.14KK763 pKa = 11.15YY764 pKa = 10.11FDD766 pKa = 4.5HH767 pKa = 7.63PDD769 pKa = 4.73FIPTYY774 pKa = 9.87KK775 pKa = 9.08VTSPVRR781 pKa = 11.84ILYY784 pKa = 7.63HH785 pKa = 5.2TLILTLKK792 pKa = 8.74LCKK795 pKa = 10.06YY796 pKa = 9.87YY797 pKa = 10.06PLFGSAVVYY806 pKa = 10.6LLLSLLMKK814 pKa = 10.29RR815 pKa = 11.84INSIMPTGPAFMKK828 pKa = 10.09PISSLFPLMAKK839 pKa = 10.26CFVTLPATFWSLGFSYY855 pKa = 10.53GFAFTYY861 pKa = 9.68VLRR864 pKa = 11.84HH865 pKa = 5.33LKK867 pKa = 10.21FRR869 pKa = 11.84TTYY872 pKa = 8.05EE873 pKa = 4.43TYY875 pKa = 10.73CKK877 pKa = 10.46VLYY880 pKa = 10.46QEE882 pKa = 5.59AEE884 pKa = 3.99GEE886 pKa = 4.12MRR888 pKa = 11.84PEE890 pKa = 4.02RR891 pKa = 11.84MHH893 pKa = 6.34HH894 pKa = 6.1QDD896 pKa = 3.83LIKK899 pKa = 11.06YY900 pKa = 9.47SDD902 pKa = 3.58PKK904 pKa = 8.45TVKK907 pKa = 10.67VKK909 pKa = 9.66VTDD912 pKa = 4.05KK913 pKa = 11.11LICQDD918 pKa = 4.58DD919 pKa = 4.05LFKK922 pKa = 10.35IDD924 pKa = 4.0EE925 pKa = 4.38EE926 pKa = 4.49VVEE929 pKa = 4.28YY930 pKa = 10.44EE931 pKa = 5.04RR932 pKa = 11.84KK933 pKa = 9.26RR934 pKa = 11.84QLNVSLEE941 pKa = 4.15LFSQFSGYY949 pKa = 8.12TMITAPTKK957 pKa = 9.33EE958 pKa = 4.49FYY960 pKa = 10.01MGLQRR965 pKa = 11.84DD966 pKa = 3.81IKK968 pKa = 8.6RR969 pKa = 11.84TCSIGFDD976 pKa = 3.34RR977 pKa = 11.84MDD979 pKa = 3.22MARR982 pKa = 11.84RR983 pKa = 11.84NIFNDD988 pKa = 2.94TATYY992 pKa = 7.46ITAWKK997 pKa = 9.43LSEE1000 pKa = 3.75MTKK1003 pKa = 10.04RR1004 pKa = 11.84DD1005 pKa = 3.42SHH1007 pKa = 5.43EE1008 pKa = 3.9QPLNN1012 pKa = 3.45
Molecular weight: 113.28 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KFP7|A0A1L3KFP7_9VIRU Uncharacterized protein OS=Beihai tombus-like virus 18 OX=1922721 PE=4 SV=1
MM1 pKa = 7.64SVINKK6 pKa = 8.82NRR8 pKa = 11.84VANITRR14 pKa = 11.84PMVDD18 pKa = 3.27RR19 pKa = 11.84DD20 pKa = 4.64DD21 pKa = 4.2IVTSATGAMHH31 pKa = 7.22RR32 pKa = 11.84VTPVMPDD39 pKa = 2.85VKK41 pKa = 10.72EE42 pKa = 3.95PEE44 pKa = 4.36EE45 pKa = 3.91YY46 pKa = 10.33RR47 pKa = 11.84KK48 pKa = 9.2FARR51 pKa = 11.84KK52 pKa = 8.28MFEE55 pKa = 3.64IFLTPLEE62 pKa = 4.0WDD64 pKa = 3.9EE65 pKa = 4.29IIKK68 pKa = 10.6FEE70 pKa = 4.91VFIDD74 pKa = 3.83MCDD77 pKa = 3.79KK78 pKa = 9.91PQSWKK83 pKa = 9.57DD84 pKa = 3.48TMRR87 pKa = 11.84EE88 pKa = 3.67CHH90 pKa = 5.73YY91 pKa = 11.31NRR93 pKa = 11.84VKK95 pKa = 11.11DD96 pKa = 3.8FDD98 pKa = 3.88YY99 pKa = 11.67EE100 pKa = 3.96EE101 pKa = 4.79FYY103 pKa = 10.37RR104 pKa = 11.84TNSSHH109 pKa = 7.05IKK111 pKa = 10.44EE112 pKa = 4.16EE113 pKa = 5.98DD114 pKa = 3.23YY115 pKa = 10.16DD116 pKa = 3.62TWKK119 pKa = 10.34NARR122 pKa = 11.84TINGTHH128 pKa = 6.56RR129 pKa = 11.84KK130 pKa = 9.96GYY132 pKa = 10.89AKK134 pKa = 10.68DD135 pKa = 3.38HH136 pKa = 6.42LSWIARR142 pKa = 11.84YY143 pKa = 9.54VKK145 pKa = 10.36SCEE148 pKa = 3.51KK149 pKa = 10.4QIYY152 pKa = 9.0HH153 pKa = 7.14KK154 pKa = 11.07LPGLVKK160 pKa = 10.63GMTPKK165 pKa = 10.64EE166 pKa = 3.95RR167 pKa = 11.84MEE169 pKa = 4.1AMYY172 pKa = 10.5EE173 pKa = 4.17LGDD176 pKa = 4.33HH177 pKa = 5.7MPKK180 pKa = 10.28TIGDD184 pKa = 3.82YY185 pKa = 10.4TSYY188 pKa = 10.53EE189 pKa = 3.74ASFKK193 pKa = 9.84RR194 pKa = 11.84KK195 pKa = 9.85KK196 pKa = 9.29MEE198 pKa = 4.02CVQFEE203 pKa = 4.35FYY205 pKa = 11.3EE206 pKa = 4.72FMFQKK211 pKa = 10.84LPFKK215 pKa = 10.34NKK217 pKa = 9.44LMEE220 pKa = 4.31GMRR223 pKa = 11.84HH224 pKa = 5.73VIGGKK229 pKa = 9.7NKK231 pKa = 8.54MRR233 pKa = 11.84FKK235 pKa = 11.06NFTWYY240 pKa = 10.58SIARR244 pKa = 11.84KK245 pKa = 8.95MSGEE249 pKa = 4.09SDD251 pKa = 3.5TALSNALDD259 pKa = 3.67NWVTWLFLLGKK270 pKa = 10.04KK271 pKa = 9.29GVPPHH276 pKa = 5.88QAAEE280 pKa = 4.11MIYY283 pKa = 10.78VEE285 pKa = 4.69GDD287 pKa = 3.53DD288 pKa = 4.98NASSLQNHH296 pKa = 6.67KK297 pKa = 10.44LDD299 pKa = 4.13KK300 pKa = 10.86KK301 pKa = 10.81DD302 pKa = 4.77FEE304 pKa = 4.99DD305 pKa = 4.4LGLKK309 pKa = 9.96IKK311 pKa = 10.55IEE313 pKa = 4.0TGLDD317 pKa = 2.98IEE319 pKa = 4.5QTGFCQLYY327 pKa = 8.09FTLPPTGTICADD339 pKa = 3.09PWKK342 pKa = 10.63KK343 pKa = 10.08LVKK346 pKa = 10.4FSKK349 pKa = 10.85APVKK353 pKa = 10.21YY354 pKa = 9.96AQASDD359 pKa = 3.58KK360 pKa = 11.0VLNSLLRR367 pKa = 11.84AQALSMLYY375 pKa = 9.51LHH377 pKa = 6.95KK378 pKa = 10.49GAPVVHH384 pKa = 6.77KK385 pKa = 10.38LAEE388 pKa = 4.05RR389 pKa = 11.84LLYY392 pKa = 8.52ITRR395 pKa = 11.84GFNVRR400 pKa = 11.84NHH402 pKa = 5.52HH403 pKa = 6.48WEE405 pKa = 3.8SAMKK409 pKa = 10.66YY410 pKa = 10.2GFDD413 pKa = 3.59GNLIKK418 pKa = 9.99TMKK421 pKa = 8.47WRR423 pKa = 11.84DD424 pKa = 3.7YY425 pKa = 10.87IDD427 pKa = 3.9TPVKK431 pKa = 10.19MEE433 pKa = 4.15DD434 pKa = 3.07RR435 pKa = 11.84VLVEE439 pKa = 4.21RR440 pKa = 11.84QFGMTIEE447 pKa = 4.03MQLLIEE453 pKa = 4.74EE454 pKa = 5.19KK455 pKa = 10.45LDD457 pKa = 3.42SWKK460 pKa = 11.03GGLLHH465 pKa = 7.1LPVDD469 pKa = 4.29WFPDD473 pKa = 3.31DD474 pKa = 3.18WALFYY479 pKa = 11.14SLYY482 pKa = 10.32ATDD485 pKa = 4.99GVHH488 pKa = 6.91SSWFSPEE495 pKa = 3.17GTLRR499 pKa = 11.84KK500 pKa = 8.89HH501 pKa = 6.43HH502 pKa = 7.33KK503 pKa = 9.5DD504 pKa = 3.06TILSFTRR511 pKa = 11.84KK512 pKa = 7.48TKK514 pKa = 10.66SGTNVLFRR522 pKa = 11.84PARR525 pKa = 11.84SPGATKK531 pKa = 10.92YY532 pKa = 10.62KK533 pKa = 10.6GFNNKK538 pKa = 9.07LSQGNN543 pKa = 3.48
MM1 pKa = 7.64SVINKK6 pKa = 8.82NRR8 pKa = 11.84VANITRR14 pKa = 11.84PMVDD18 pKa = 3.27RR19 pKa = 11.84DD20 pKa = 4.64DD21 pKa = 4.2IVTSATGAMHH31 pKa = 7.22RR32 pKa = 11.84VTPVMPDD39 pKa = 2.85VKK41 pKa = 10.72EE42 pKa = 3.95PEE44 pKa = 4.36EE45 pKa = 3.91YY46 pKa = 10.33RR47 pKa = 11.84KK48 pKa = 9.2FARR51 pKa = 11.84KK52 pKa = 8.28MFEE55 pKa = 3.64IFLTPLEE62 pKa = 4.0WDD64 pKa = 3.9EE65 pKa = 4.29IIKK68 pKa = 10.6FEE70 pKa = 4.91VFIDD74 pKa = 3.83MCDD77 pKa = 3.79KK78 pKa = 9.91PQSWKK83 pKa = 9.57DD84 pKa = 3.48TMRR87 pKa = 11.84EE88 pKa = 3.67CHH90 pKa = 5.73YY91 pKa = 11.31NRR93 pKa = 11.84VKK95 pKa = 11.11DD96 pKa = 3.8FDD98 pKa = 3.88YY99 pKa = 11.67EE100 pKa = 3.96EE101 pKa = 4.79FYY103 pKa = 10.37RR104 pKa = 11.84TNSSHH109 pKa = 7.05IKK111 pKa = 10.44EE112 pKa = 4.16EE113 pKa = 5.98DD114 pKa = 3.23YY115 pKa = 10.16DD116 pKa = 3.62TWKK119 pKa = 10.34NARR122 pKa = 11.84TINGTHH128 pKa = 6.56RR129 pKa = 11.84KK130 pKa = 9.96GYY132 pKa = 10.89AKK134 pKa = 10.68DD135 pKa = 3.38HH136 pKa = 6.42LSWIARR142 pKa = 11.84YY143 pKa = 9.54VKK145 pKa = 10.36SCEE148 pKa = 3.51KK149 pKa = 10.4QIYY152 pKa = 9.0HH153 pKa = 7.14KK154 pKa = 11.07LPGLVKK160 pKa = 10.63GMTPKK165 pKa = 10.64EE166 pKa = 3.95RR167 pKa = 11.84MEE169 pKa = 4.1AMYY172 pKa = 10.5EE173 pKa = 4.17LGDD176 pKa = 4.33HH177 pKa = 5.7MPKK180 pKa = 10.28TIGDD184 pKa = 3.82YY185 pKa = 10.4TSYY188 pKa = 10.53EE189 pKa = 3.74ASFKK193 pKa = 9.84RR194 pKa = 11.84KK195 pKa = 9.85KK196 pKa = 9.29MEE198 pKa = 4.02CVQFEE203 pKa = 4.35FYY205 pKa = 11.3EE206 pKa = 4.72FMFQKK211 pKa = 10.84LPFKK215 pKa = 10.34NKK217 pKa = 9.44LMEE220 pKa = 4.31GMRR223 pKa = 11.84HH224 pKa = 5.73VIGGKK229 pKa = 9.7NKK231 pKa = 8.54MRR233 pKa = 11.84FKK235 pKa = 11.06NFTWYY240 pKa = 10.58SIARR244 pKa = 11.84KK245 pKa = 8.95MSGEE249 pKa = 4.09SDD251 pKa = 3.5TALSNALDD259 pKa = 3.67NWVTWLFLLGKK270 pKa = 10.04KK271 pKa = 9.29GVPPHH276 pKa = 5.88QAAEE280 pKa = 4.11MIYY283 pKa = 10.78VEE285 pKa = 4.69GDD287 pKa = 3.53DD288 pKa = 4.98NASSLQNHH296 pKa = 6.67KK297 pKa = 10.44LDD299 pKa = 4.13KK300 pKa = 10.86KK301 pKa = 10.81DD302 pKa = 4.77FEE304 pKa = 4.99DD305 pKa = 4.4LGLKK309 pKa = 9.96IKK311 pKa = 10.55IEE313 pKa = 4.0TGLDD317 pKa = 2.98IEE319 pKa = 4.5QTGFCQLYY327 pKa = 8.09FTLPPTGTICADD339 pKa = 3.09PWKK342 pKa = 10.63KK343 pKa = 10.08LVKK346 pKa = 10.4FSKK349 pKa = 10.85APVKK353 pKa = 10.21YY354 pKa = 9.96AQASDD359 pKa = 3.58KK360 pKa = 11.0VLNSLLRR367 pKa = 11.84AQALSMLYY375 pKa = 9.51LHH377 pKa = 6.95KK378 pKa = 10.49GAPVVHH384 pKa = 6.77KK385 pKa = 10.38LAEE388 pKa = 4.05RR389 pKa = 11.84LLYY392 pKa = 8.52ITRR395 pKa = 11.84GFNVRR400 pKa = 11.84NHH402 pKa = 5.52HH403 pKa = 6.48WEE405 pKa = 3.8SAMKK409 pKa = 10.66YY410 pKa = 10.2GFDD413 pKa = 3.59GNLIKK418 pKa = 9.99TMKK421 pKa = 8.47WRR423 pKa = 11.84DD424 pKa = 3.7YY425 pKa = 10.87IDD427 pKa = 3.9TPVKK431 pKa = 10.19MEE433 pKa = 4.15DD434 pKa = 3.07RR435 pKa = 11.84VLVEE439 pKa = 4.21RR440 pKa = 11.84QFGMTIEE447 pKa = 4.03MQLLIEE453 pKa = 4.74EE454 pKa = 5.19KK455 pKa = 10.45LDD457 pKa = 3.42SWKK460 pKa = 11.03GGLLHH465 pKa = 7.1LPVDD469 pKa = 4.29WFPDD473 pKa = 3.31DD474 pKa = 3.18WALFYY479 pKa = 11.14SLYY482 pKa = 10.32ATDD485 pKa = 4.99GVHH488 pKa = 6.91SSWFSPEE495 pKa = 3.17GTLRR499 pKa = 11.84KK500 pKa = 8.89HH501 pKa = 6.43HH502 pKa = 7.33KK503 pKa = 9.5DD504 pKa = 3.06TILSFTRR511 pKa = 11.84KK512 pKa = 7.48TKK514 pKa = 10.66SGTNVLFRR522 pKa = 11.84PARR525 pKa = 11.84SPGATKK531 pKa = 10.92YY532 pKa = 10.62KK533 pKa = 10.6GFNNKK538 pKa = 9.07LSQGNN543 pKa = 3.48
Molecular weight: 63.49 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1555 |
543 |
1012 |
777.5 |
88.39 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.688 ± 0.865 | 1.35 ± 0.139 |
6.109 ± 0.19 | 5.852 ± 0.335 |
5.145 ± 0.111 | 6.045 ± 0.19 |
2.444 ± 0.492 | 4.952 ± 0.196 |
7.524 ± 1.575 | 7.524 ± 0.327 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.151 ± 0.716 | 4.695 ± 0.363 |
5.08 ± 0.477 | 3.215 ± 0.464 |
5.402 ± 0.139 | 5.723 ± 0.216 |
6.881 ± 0.558 | 6.817 ± 0.937 |
1.736 ± 0.475 | 3.601 ± 0.358 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
