
Wuhan spider virus 4
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.75
Get precalculated fractions of proteins
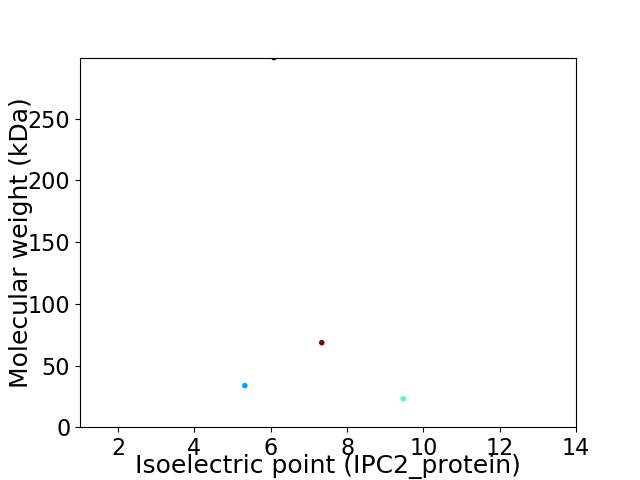
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
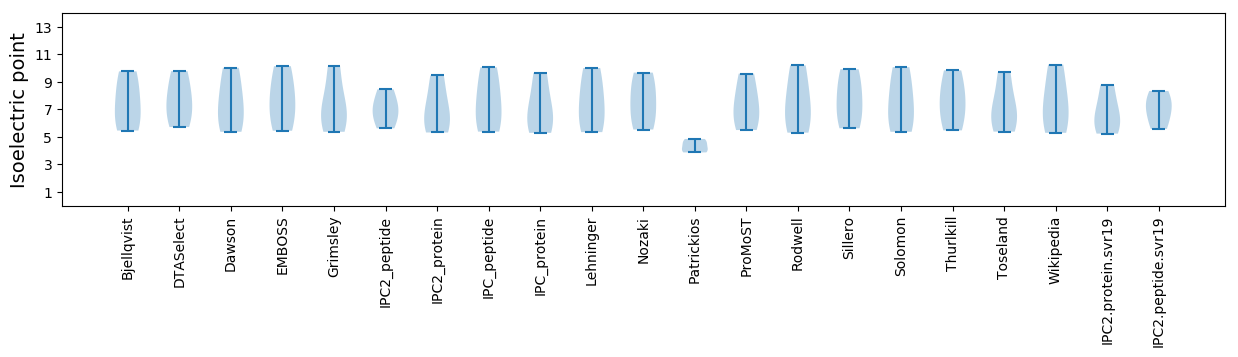
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KJ44|A0A1L3KJ44_9VIRU Uncharacterized protein OS=Wuhan spider virus 4 OX=1923753 PE=4 SV=1
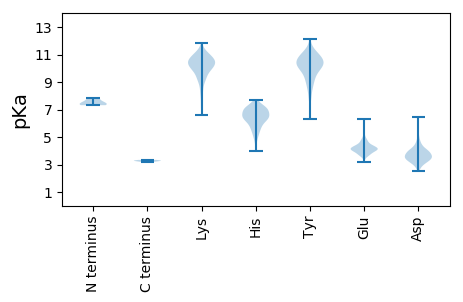
MM1 pKa = 7.5SKK3 pKa = 9.48MGAEE7 pKa = 4.59CSNAPAGDD15 pKa = 3.66CQFYY19 pKa = 10.77KK20 pKa = 10.99GVLNTYY26 pKa = 9.96AQIIRR31 pKa = 11.84QILEE35 pKa = 3.51QLEE38 pKa = 4.33AYY40 pKa = 9.9KK41 pKa = 10.63DD42 pKa = 4.0FPYY45 pKa = 10.5KK46 pKa = 10.97ANDD49 pKa = 3.62YY50 pKa = 10.55YY51 pKa = 12.16VMLTQVIISASDD63 pKa = 3.51DD64 pKa = 4.22DD65 pKa = 4.96FDD67 pKa = 4.16NCRR70 pKa = 11.84ALLASMDD77 pKa = 3.38SWIEE81 pKa = 3.89EE82 pKa = 3.72ISPFDD87 pKa = 3.53ARR89 pKa = 11.84DD90 pKa = 3.66VISRR94 pKa = 11.84TLMQALSKK102 pKa = 10.25FHH104 pKa = 7.24FSFKK108 pKa = 10.79QNLIDD113 pKa = 4.14NLKK116 pKa = 9.66KK117 pKa = 10.52ACPKK121 pKa = 10.09KK122 pKa = 10.78VRR124 pKa = 11.84TPLHH128 pKa = 5.65SQCSSCDD135 pKa = 3.38HH136 pKa = 6.4FPEE139 pKa = 5.75KK140 pKa = 10.61SEE142 pKa = 4.24EE143 pKa = 4.03EE144 pKa = 4.41VEE146 pKa = 4.5VVRR149 pKa = 11.84NAIYY153 pKa = 10.66EE154 pKa = 4.28MYY156 pKa = 10.92LEE158 pKa = 4.74DD159 pKa = 4.07QDD161 pKa = 3.95LKK163 pKa = 11.18RR164 pKa = 11.84RR165 pKa = 11.84GVLTRR170 pKa = 11.84KK171 pKa = 9.54PPSHH175 pKa = 6.65SFGGEE180 pKa = 4.04TIFKK184 pKa = 9.07PVPIPAPVHH193 pKa = 5.75KK194 pKa = 10.21SPPKK198 pKa = 8.05TPEE201 pKa = 3.63RR202 pKa = 11.84RR203 pKa = 11.84PSRR206 pKa = 11.84PSSRR210 pKa = 11.84NSQKK214 pKa = 10.46VYY216 pKa = 10.51RR217 pKa = 11.84CDD219 pKa = 3.25IPCYY223 pKa = 8.15PTQEE227 pKa = 4.27EE228 pKa = 4.2ANRR231 pKa = 11.84AYY233 pKa = 10.63RR234 pKa = 11.84KK235 pKa = 9.12AQHH238 pKa = 6.64DD239 pKa = 3.18AWFYY243 pKa = 11.21KK244 pKa = 9.98HH245 pKa = 6.43EE246 pKa = 4.16EE247 pKa = 3.99WPMEE251 pKa = 3.66ICEE254 pKa = 4.15PQIGEE259 pKa = 4.25PEE261 pKa = 4.46VYY263 pKa = 10.2DD264 pKa = 4.79QEE266 pKa = 5.57QDD268 pKa = 4.21DD269 pKa = 4.04SWRR272 pKa = 11.84DD273 pKa = 3.32PNVYY277 pKa = 10.52DD278 pKa = 5.25DD279 pKa = 5.64DD280 pKa = 4.13GTPFHH285 pKa = 6.96PRR287 pKa = 11.84NLPP290 pKa = 3.34
MM1 pKa = 7.5SKK3 pKa = 9.48MGAEE7 pKa = 4.59CSNAPAGDD15 pKa = 3.66CQFYY19 pKa = 10.77KK20 pKa = 10.99GVLNTYY26 pKa = 9.96AQIIRR31 pKa = 11.84QILEE35 pKa = 3.51QLEE38 pKa = 4.33AYY40 pKa = 9.9KK41 pKa = 10.63DD42 pKa = 4.0FPYY45 pKa = 10.5KK46 pKa = 10.97ANDD49 pKa = 3.62YY50 pKa = 10.55YY51 pKa = 12.16VMLTQVIISASDD63 pKa = 3.51DD64 pKa = 4.22DD65 pKa = 4.96FDD67 pKa = 4.16NCRR70 pKa = 11.84ALLASMDD77 pKa = 3.38SWIEE81 pKa = 3.89EE82 pKa = 3.72ISPFDD87 pKa = 3.53ARR89 pKa = 11.84DD90 pKa = 3.66VISRR94 pKa = 11.84TLMQALSKK102 pKa = 10.25FHH104 pKa = 7.24FSFKK108 pKa = 10.79QNLIDD113 pKa = 4.14NLKK116 pKa = 9.66KK117 pKa = 10.52ACPKK121 pKa = 10.09KK122 pKa = 10.78VRR124 pKa = 11.84TPLHH128 pKa = 5.65SQCSSCDD135 pKa = 3.38HH136 pKa = 6.4FPEE139 pKa = 5.75KK140 pKa = 10.61SEE142 pKa = 4.24EE143 pKa = 4.03EE144 pKa = 4.41VEE146 pKa = 4.5VVRR149 pKa = 11.84NAIYY153 pKa = 10.66EE154 pKa = 4.28MYY156 pKa = 10.92LEE158 pKa = 4.74DD159 pKa = 4.07QDD161 pKa = 3.95LKK163 pKa = 11.18RR164 pKa = 11.84RR165 pKa = 11.84GVLTRR170 pKa = 11.84KK171 pKa = 9.54PPSHH175 pKa = 6.65SFGGEE180 pKa = 4.04TIFKK184 pKa = 9.07PVPIPAPVHH193 pKa = 5.75KK194 pKa = 10.21SPPKK198 pKa = 8.05TPEE201 pKa = 3.63RR202 pKa = 11.84RR203 pKa = 11.84PSRR206 pKa = 11.84PSSRR210 pKa = 11.84NSQKK214 pKa = 10.46VYY216 pKa = 10.51RR217 pKa = 11.84CDD219 pKa = 3.25IPCYY223 pKa = 8.15PTQEE227 pKa = 4.27EE228 pKa = 4.2ANRR231 pKa = 11.84AYY233 pKa = 10.63RR234 pKa = 11.84KK235 pKa = 9.12AQHH238 pKa = 6.64DD239 pKa = 3.18AWFYY243 pKa = 11.21KK244 pKa = 9.98HH245 pKa = 6.43EE246 pKa = 4.16EE247 pKa = 3.99WPMEE251 pKa = 3.66ICEE254 pKa = 4.15PQIGEE259 pKa = 4.25PEE261 pKa = 4.46VYY263 pKa = 10.2DD264 pKa = 4.79QEE266 pKa = 5.57QDD268 pKa = 4.21DD269 pKa = 4.04SWRR272 pKa = 11.84DD273 pKa = 3.32PNVYY277 pKa = 10.52DD278 pKa = 5.25DD279 pKa = 5.64DD280 pKa = 4.13GTPFHH285 pKa = 6.96PRR287 pKa = 11.84NLPP290 pKa = 3.34
Molecular weight: 33.67 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KJB8|A0A1L3KJB8_9VIRU Calici_coat domain-containing protein OS=Wuhan spider virus 4 OX=1923753 PE=4 SV=1
MM1 pKa = 7.37AAAALAAAVGGSTISAGATLGASLGAAQIQRR32 pKa = 11.84NTSLQVQDD40 pKa = 3.87SQQKK44 pKa = 7.33FTEE47 pKa = 4.89GIINRR52 pKa = 11.84GEE54 pKa = 3.81QAFTKK59 pKa = 10.47NGLPGYY65 pKa = 9.49LYY67 pKa = 9.86WQGTGPMPNTLYY79 pKa = 10.47QVNGANFAEE88 pKa = 4.74GFGVNTNLPYY98 pKa = 9.58YY99 pKa = 8.99TSNPMSQIYY108 pKa = 9.84KK109 pKa = 10.01AGKK112 pKa = 7.62PQYY115 pKa = 9.45SNKK118 pKa = 10.1DD119 pKa = 3.28ATEE122 pKa = 4.09TPPTKK127 pKa = 9.91ISSSPQSNTQPKK139 pKa = 8.25TFEE142 pKa = 4.41PPSEE146 pKa = 4.15FNRR149 pKa = 11.84TGWQSDD155 pKa = 3.44RR156 pKa = 11.84AGLGQGRR163 pKa = 11.84FASNYY168 pKa = 9.24NAVPPPNFNSAATQTRR184 pKa = 11.84NDD186 pKa = 3.35ALIRR190 pKa = 11.84SVGVQFPGPGPRR202 pKa = 11.84IITPRR207 pKa = 11.84VPSSYY212 pKa = 10.47RR213 pKa = 11.84AINN216 pKa = 3.2
MM1 pKa = 7.37AAAALAAAVGGSTISAGATLGASLGAAQIQRR32 pKa = 11.84NTSLQVQDD40 pKa = 3.87SQQKK44 pKa = 7.33FTEE47 pKa = 4.89GIINRR52 pKa = 11.84GEE54 pKa = 3.81QAFTKK59 pKa = 10.47NGLPGYY65 pKa = 9.49LYY67 pKa = 9.86WQGTGPMPNTLYY79 pKa = 10.47QVNGANFAEE88 pKa = 4.74GFGVNTNLPYY98 pKa = 9.58YY99 pKa = 8.99TSNPMSQIYY108 pKa = 9.84KK109 pKa = 10.01AGKK112 pKa = 7.62PQYY115 pKa = 9.45SNKK118 pKa = 10.1DD119 pKa = 3.28ATEE122 pKa = 4.09TPPTKK127 pKa = 9.91ISSSPQSNTQPKK139 pKa = 8.25TFEE142 pKa = 4.41PPSEE146 pKa = 4.15FNRR149 pKa = 11.84TGWQSDD155 pKa = 3.44RR156 pKa = 11.84AGLGQGRR163 pKa = 11.84FASNYY168 pKa = 9.24NAVPPPNFNSAATQTRR184 pKa = 11.84NDD186 pKa = 3.35ALIRR190 pKa = 11.84SVGVQFPGPGPRR202 pKa = 11.84IITPRR207 pKa = 11.84VPSSYY212 pKa = 10.47RR213 pKa = 11.84AINN216 pKa = 3.2
Molecular weight: 22.94 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3749 |
216 |
2628 |
937.3 |
106.16 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.428 ± 0.738 | 1.867 ± 0.531 |
5.521 ± 0.601 | 6.082 ± 1.002 |
4.615 ± 0.535 | 5.361 ± 0.881 |
2.214 ± 0.395 | 5.788 ± 0.63 |
6.722 ± 1.047 | 7.709 ± 0.803 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.107 ± 0.136 | 5.308 ± 0.55 |
5.628 ± 1.485 | 4.481 ± 0.702 |
4.428 ± 0.359 | 7.495 ± 0.663 |
5.601 ± 0.496 | 6.828 ± 0.658 |
1.654 ± 0.155 | 4.161 ± 0.547 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
